Table of contents
- 1. Introduction to Biology2h 42m
- 2. Chemistry3h 40m
- 3. Water1h 26m
- 4. Biomolecules2h 23m
- 5. Cell Components2h 26m
- 6. The Membrane2h 31m
- 7. Energy and Metabolism2h 0m
- 8. Respiration2h 40m
- 9. Photosynthesis2h 49m
- 10. Cell Signaling59m
- 11. Cell Division2h 47m
- 12. Meiosis2h 0m
- 13. Mendelian Genetics4h 44m
- Introduction to Mendel's Experiments7m
- Genotype vs. Phenotype17m
- Punnett Squares13m
- Mendel's Experiments26m
- Mendel's Laws18m
- Monohybrid Crosses19m
- Test Crosses14m
- Dihybrid Crosses20m
- Punnett Square Probability26m
- Incomplete Dominance vs. Codominance20m
- Epistasis7m
- Non-Mendelian Genetics12m
- Pedigrees6m
- Autosomal Inheritance21m
- Sex-Linked Inheritance43m
- X-Inactivation9m
- 14. DNA Synthesis2h 27m
- 15. Gene Expression3h 20m
- 16. Regulation of Expression3h 31m
- Introduction to Regulation of Gene Expression13m
- Prokaryotic Gene Regulation via Operons27m
- The Lac Operon21m
- Glucose's Impact on Lac Operon25m
- The Trp Operon20m
- Review of the Lac Operon & Trp Operon11m
- Introduction to Eukaryotic Gene Regulation9m
- Eukaryotic Chromatin Modifications16m
- Eukaryotic Transcriptional Control22m
- Eukaryotic Post-Transcriptional Regulation28m
- Eukaryotic Post-Translational Regulation13m
- 17. Viruses37m
- 18. Biotechnology2h 58m
- 19. Genomics17m
- 20. Development1h 5m
- 21. Evolution3h 1m
- 22. Evolution of Populations3h 53m
- 23. Speciation1h 37m
- 24. History of Life on Earth2h 6m
- 25. Phylogeny2h 31m
- 26. Prokaryotes4h 59m
- 27. Protists1h 12m
- 28. Plants1h 22m
- 29. Fungi36m
- 30. Overview of Animals34m
- 31. Invertebrates1h 2m
- 32. Vertebrates50m
- 33. Plant Anatomy1h 3m
- 34. Vascular Plant Transport1h 2m
- 35. Soil37m
- 36. Plant Reproduction47m
- 37. Plant Sensation and Response1h 9m
- 38. Animal Form and Function1h 19m
- 39. Digestive System1h 10m
- 40. Circulatory System1h 49m
- 41. Immune System1h 12m
- 42. Osmoregulation and Excretion50m
- 43. Endocrine System1h 4m
- 44. Animal Reproduction1h 2m
- 45. Nervous System1h 55m
- 46. Sensory Systems46m
- 47. Muscle Systems23m
- 48. Ecology3h 11m
- Introduction to Ecology20m
- Biogeography14m
- Earth's Climate Patterns50m
- Introduction to Terrestrial Biomes10m
- Terrestrial Biomes: Near Equator13m
- Terrestrial Biomes: Temperate Regions10m
- Terrestrial Biomes: Northern Regions15m
- Introduction to Aquatic Biomes27m
- Freshwater Aquatic Biomes14m
- Marine Aquatic Biomes13m
- 49. Animal Behavior28m
- 50. Population Ecology3h 41m
- Introduction to Population Ecology28m
- Population Sampling Methods23m
- Life History12m
- Population Demography17m
- Factors Limiting Population Growth14m
- Introduction to Population Growth Models22m
- Linear Population Growth6m
- Exponential Population Growth29m
- Logistic Population Growth32m
- r/K Selection10m
- The Human Population22m
- 51. Community Ecology2h 46m
- Introduction to Community Ecology2m
- Introduction to Community Interactions9m
- Community Interactions: Competition (-/-)38m
- Community Interactions: Exploitation (+/-)23m
- Community Interactions: Mutualism (+/+) & Commensalism (+/0)9m
- Community Structure35m
- Community Dynamics26m
- Geographic Impact on Communities21m
- 52. Ecosystems2h 36m
- 53. Conservation Biology24m
22. Evolution of Populations
The Hardy-Weinberg Principle
Problem 4
Textbook Question
There are 25 individuals in population 1, all with genotype AA, and there are 40 individuals in population 2, all with genotype aa. Assume that these populations are located far from each other and that their environmental conditions are very similar. Based on the information given here, the observed genetic variation most likely resulted from
a. Genetic drift.
b. Gene flow.
c. Nonrandom mating.
d. Directional selection.
 Verified step by step guidance
Verified step by step guidance1
Understand the genotypes present in each population: Population 1 consists entirely of individuals with genotype AA, while Population 2 consists entirely of individuals with genotype aa.
Consider the concept of genetic drift: Genetic drift refers to random changes in allele frequencies in a population, which is more pronounced in small populations. However, since both populations are fixed for different alleles (AA and aa), genetic drift is unlikely to be the cause of the observed genetic variation.
Evaluate the possibility of gene flow: Gene flow involves the transfer of alleles between populations. Given that the populations are located far from each other, gene flow is unlikely to occur, as there is no exchange of individuals or alleles between the populations.
Analyze the role of nonrandom mating: Nonrandom mating affects genotype frequencies but not allele frequencies. Since both populations are fixed for different alleles, nonrandom mating is not a contributing factor to the observed genetic variation.
Consider directional selection: Directional selection favors one allele over another, leading to changes in allele frequencies. However, since both populations are fixed for different alleles and the environmental conditions are similar, directional selection is unlikely to be responsible for the observed genetic variation.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
3mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Genetic Drift
Genetic drift refers to random changes in allele frequencies within a population, often having a more pronounced effect in small populations. It can lead to the loss or fixation of alleles over time, independent of natural selection. In isolated populations, genetic drift can significantly impact genetic variation, especially when population sizes are small.
Recommended video:

Genetic Drift
Gene Flow
Gene flow is the transfer of genetic material between separate populations, often through migration. It can introduce new alleles into a population, increasing genetic diversity and potentially altering allele frequencies. In the context of isolated populations, gene flow is unlikely unless individuals from different populations interbreed, which is not the case here.
Recommended video:
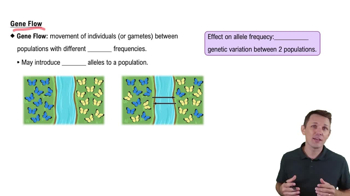
Gene Flow
Directional Selection
Directional selection is a form of natural selection where one extreme phenotype is favored over others, leading to a shift in allele frequencies in a particular direction. This process can result in the predominance of certain traits within a population. However, in the given scenario, the environmental conditions are similar, making directional selection an unlikely cause of genetic variation.
Recommended video:

Natural Selection

 6:29m
6:29mWatch next
Master Introduction to Hardy-Weinberg with a bite sized video explanation from Jason
Start learningRelated Videos
Related Practice
Multiple Choice
Consider a gene that exists in two allelic forms in a simple Mendelian dominant/recessive pair. In a large population of randomly breeding organisms, the frequency of a recessive allele is initially 0.3. There is no migration and no selection. Humans enter this ecosystem and selectively hunt individuals showing the dominant trait. When the gene frequency is reexamined at the end of the year, __________.
1275
views
1
rank
