Suppose you have a 1-kb segment of cloned DNA that is suspected to contain a eukaryotic promoter, including a TATA box, a CAAT box, and an upstream GC-rich sequence. The clone also contains a gene whose transcript is readily detectable. Your laboratory supervisor asks you to outline an experiment that will (1) determine if eukaryotic transcription factors (TF) bind to the fragment and, if so, (2) identify where on the fragment the transcription factors bind. All necessary reagents, equipment, and experimental know-how are available in the laboratory. Your assignment is to propose techniques to be used to address the two items your supervisor has listed and to describe the kind of results that would indicate binding of TF to the DNA and the location of the binding.
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
18. Molecular Genetic Tools
Methods for Analyzing DNA
Problem 29
Textbook Question
The following dideoxy DNA sequencing gel is produced in a laboratory.
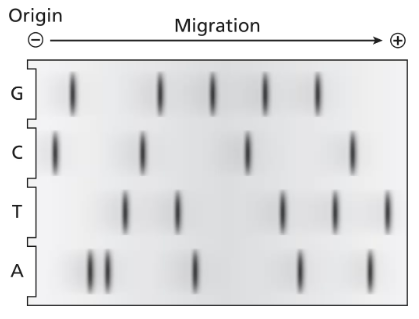
What is the double-stranded DNA sequence of this molecule? Label the polarity of each strand.
 Verified step by step guidance
Verified step by step guidance1
Examine the dideoxy DNA sequencing gel. Identify the lanes corresponding to the four dideoxynucleotides (ddATP, ddTTP, ddCTP, ddGTP), which terminate DNA synthesis at specific bases (A, T, C, G).
Read the sequence from the bottom of the gel to the top, as the smallest fragments (closest to the primer) migrate the furthest. This sequence represents the newly synthesized strand in the 5' to 3' direction.
Determine the complementary strand by pairing the bases of the synthesized strand with their complementary bases (A pairs with T, and C pairs with G). Write this complementary strand in the 3' to 5' direction.
Label the polarity of both strands. The synthesized strand is 5' to 3', and the complementary strand is 3' to 5'.
Combine the information to write the double-stranded DNA sequence, ensuring that the polarity of each strand is clearly labeled (e.g., 5' to 3' for one strand and 3' to 5' for the complementary strand).
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
2mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Dideoxy DNA Sequencing
Dideoxy DNA sequencing, also known as Sanger sequencing, is a method used to determine the nucleotide sequence of DNA. It involves the incorporation of dideoxynucleotides, which terminate DNA strand elongation during replication. This results in fragments of varying lengths that can be separated by gel electrophoresis, allowing for the identification of the sequence based on the size of the fragments.
Recommended video:
Guided course

Sequencing Difficulties
Polarity of DNA Strands
DNA strands have polarity, indicated as 5' to 3' and 3' to 5'. The 5' end has a phosphate group, while the 3' end has a hydroxyl group. When determining the sequence from a gel, it is essential to label the strands correctly, as the 5' to 3' directionality affects how the DNA is synthesized and read during replication and sequencing.
Recommended video:
Guided course

Double Strand Breaks
Gel Electrophoresis
Gel electrophoresis is a technique used to separate DNA fragments based on their size. In the context of dideoxy sequencing, the DNA fragments produced during the sequencing reaction are loaded into a gel matrix and subjected to an electric field. Smaller fragments migrate faster than larger ones, allowing for the visualization of the sequence by comparing the positions of the bands on the gel.
Recommended video:
Guided course

Proteomics

 7:40m
7:40mWatch next
Master Methods for Analyzing DNA and RNA with a bite sized video explanation from Kylia
Start learningRelated Videos
Related Practice
Textbook Question
407
views
