How would you design a genetic screen to find genes involved in meiosis?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
15. Genomes and Genomics
Functional Genomics
Problem 25b
Textbook Question
In conducting the study described in Problem 24, you have noted that a set of S. cerevisiae genes are repressed when yeast are grown under high-salt conditions. How might you approach this question if genome sequences for the related Saccharomyces species S. paradoxus, S. mikatae, and S. bayanus were also available?
 Verified step by step guidance
Verified step by step guidance1
Step 1: Begin by identifying the genes in S. cerevisiae that are repressed under high-salt conditions. Use transcriptomic data or gene expression profiling to pinpoint these genes.
Step 2: Obtain the genome sequences for the related Saccharomyces species (S. paradoxus, S. mikatae, and S. bayanus). Ensure the sequences are properly annotated to facilitate comparative analysis.
Step 3: Perform a comparative genomic analysis to identify orthologous genes in the related species. Use sequence alignment tools such as BLAST or Clustal Omega to find homologous genes across the species.
Step 4: Analyze the regulatory regions (promoters and enhancers) of the orthologous genes in all species. Look for conserved motifs or transcription factor binding sites that might be involved in salt-induced repression. Tools like MEME or FIMO can help identify these motifs.
Step 5: Investigate whether the repression mechanism is conserved across species by conducting functional assays, such as reporter gene experiments or mutational analysis, to test the activity of regulatory elements under high-salt conditions.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
2mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Gene Regulation
Gene regulation refers to the mechanisms that control the expression of genes, determining when and how much of a gene product is produced. In the context of S. cerevisiae, understanding how high-salt conditions repress certain genes is crucial for exploring adaptive responses and metabolic pathways. This concept is fundamental for analyzing the effects of environmental stressors on gene expression.
Recommended video:
Guided course

Review of Regulation
Comparative Genomics
Comparative genomics involves comparing the genomic features of different organisms to understand evolutionary relationships and functional similarities. By examining the genome sequences of S. paradoxus, S. mikatae, and S. bayanus alongside S. cerevisiae, researchers can identify conserved regulatory elements and gene functions that may contribute to salt tolerance or stress responses across species.
Recommended video:
Guided course

Genomics Overview
Phylogenetic Analysis
Phylogenetic analysis is the study of evolutionary relationships among biological entities, often using genetic data to construct evolutionary trees. In this scenario, analyzing the phylogenetic relationships among the Saccharomyces species can provide insights into how gene repression mechanisms have evolved in response to high-salt environments, revealing potential adaptive strategies that may differ among species.
Recommended video:
Guided course
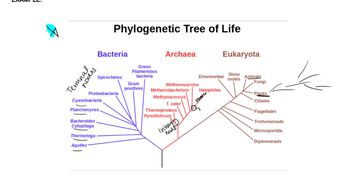
Phylogenetic Trees
Related Videos
Related Practice
Textbook Question
472
views


