How would the results vary in cross (a) of Problem 32 if genes A and B were linked with no crossing over between them? How would the results of cross (a) vary if genes A and B were linked and 20 map units (mu) apart?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
4. Genetic Mapping and Linkage
Mapping Genes
Problem 35a
Textbook Question
Based on previous family studies, an autosomal recessive disease with alleles A and a is suspected to be linked to an RFLP marker. The RFLP marker has four alleles, R₁, R₂, R₃, and R₄. The accompanying pedigree shows a three-generation family in which the disease is present. The gel shows the RFLP alleles for each family member directly below the pedigree symbol for that person. After determining the genotypes for the RFLP and disease gene for each family member, answer the following questions.
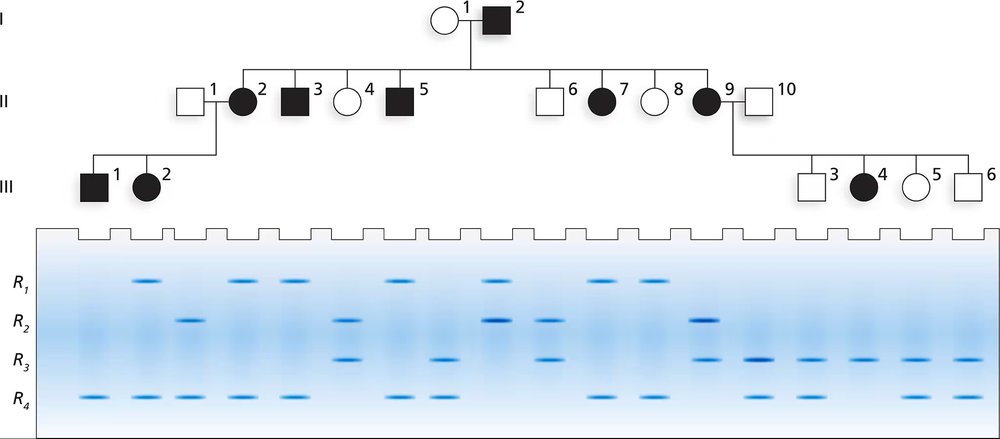
What is the most likely arrangement of syntenic alleles for the RFLP and the disease gene in I-1 and I-2?
 Verified step by step guidance
Verified step by step guidance1
Step 1: Understand the problem. The question involves determining the arrangement of syntenic alleles (alleles located on the same chromosome) for an autosomal recessive disease gene (A/a) and an RFLP marker with four alleles (R₁, R₂, R₃, R₄). The goal is to analyze the pedigree and gel electrophoresis data to deduce the most likely arrangement of these alleles in individuals I-1 and I-2.
Step 2: Analyze the inheritance pattern of the disease. Since the disease is autosomal recessive, affected individuals must have the genotype 'aa'. Unaffected individuals can be either 'AA' or 'Aa'. Use this information to assign possible genotypes for the disease gene to each family member in the pedigree.
Step 3: Examine the gel electrophoresis data for the RFLP marker. The gel shows the RFLP alleles for each individual. Assign the RFLP genotypes (e.g., R₁R₂, R₃R₄, etc.) to each family member based on the bands visible in the gel.
Step 4: Correlate the disease gene alleles with the RFLP marker alleles. Look for patterns of co-segregation between the disease gene (A/a) and specific RFLP alleles (R₁, R₂, R₃, R₄) across the family. This will help identify which RFLP alleles are most likely linked to the 'a' allele of the disease gene in individuals I-1 and I-2.
Step 5: Deduce the most likely arrangement of syntenic alleles for I-1 and I-2. Based on the co-segregation analysis, determine which RFLP alleles are linked to the 'A' and 'a' alleles in these individuals. For example, if R₁ is consistently inherited with 'a', then the arrangement for I-1 might be A-R₂/a-R₁. Repeat this process for I-2.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
4mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Autosomal Recessive Inheritance
Autosomal recessive inheritance occurs when two copies of a mutated gene are required for an individual to express a trait or disease. In this case, alleles A (normal) and a (mutated) determine the presence of the disease. Individuals with the genotype aa will exhibit the disease, while those with AA or Aa will be unaffected. Understanding this inheritance pattern is crucial for analyzing the pedigree and determining the genotypes of family members.
Recommended video:
Guided course

Autosomal Pedigrees
Restriction Fragment Length Polymorphism (RFLP)
RFLP is a molecular technique used to analyze the variations in DNA sequences. It involves digesting DNA with specific restriction enzymes, resulting in fragments of different lengths that can be separated by gel electrophoresis. The presence of different alleles (R₁, R₂, R₃, R₄) at the RFLP marker can help establish genetic linkage to the disease gene, providing insights into inheritance patterns and potential carrier status within the family.
Recommended video:
Guided course

Mapping with Markers
Syntenic Alleles
Syntenic alleles refer to genes or genetic markers that are located on the same chromosome and are inherited together due to their proximity. In the context of the question, determining the arrangement of syntenic alleles for the RFLP and disease gene in individuals I-1 and I-2 involves analyzing their genotypes to see how the alleles are linked. This understanding is essential for predicting inheritance patterns and assessing the likelihood of disease expression in future generations.
Recommended video:
Guided course

New Alleles and Migration
Related Videos
Related Practice
Textbook Question
740
views


