Synteny describes conservation of what?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
15. Genomes and Genomics
Comparative Genomics
Problem 6
Textbook Question
Annotation involves identifying genes and gene-regulatory sequences in a genome. List and describe characteristics of a genome that are hallmarks for identifying genes in an unknown sequence. What characteristics would you look for in a bacterial genome? A eukaryotic genome?
 Verified step by step guidance
Verified step by step guidance1
Understand that gene annotation involves recognizing specific features in DNA sequences that indicate the presence of genes or regulatory elements. These features differ between bacterial and eukaryotic genomes due to their structural and functional differences.
For a bacterial genome, look for characteristics such as: a) Open Reading Frames (ORFs) that are long stretches without stop codons, b) Promoter sequences like the -10 (Pribnow box) and -35 regions upstream of genes, c) Ribosome Binding Sites (Shine-Dalgarno sequences) near the start codon, d) Operon structures where multiple genes are transcribed together, and e) Relatively compact genomes with little non-coding DNA.
For a eukaryotic genome, identify features such as: a) Exons and introns within genes, requiring detection of splice sites (donor and acceptor sites), b) Promoter regions including TATA boxes and other regulatory elements upstream of transcription start sites, c) Polyadenylation signals downstream of coding regions, d) Larger intergenic regions and repetitive sequences, and e) Presence of untranslated regions (UTRs) at the 5' and 3' ends of transcripts.
Use computational tools to scan the sequence for these hallmarks: for example, algorithms that detect ORFs, promoter motifs, splice sites, and conserved regulatory sequences. Comparative genomics can also help by identifying conserved sequences across related species.
Summarize that bacterial genomes are generally simpler with continuous coding sequences and clear promoter and ribosome binding sites, while eukaryotic genomes require more complex analysis due to introns, extensive regulatory regions, and larger non-coding DNA.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
56sPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Gene Structure and Regulatory Elements
Genes consist of coding regions (exons) and non-coding regions (introns in eukaryotes), along with regulatory sequences like promoters and enhancers. Identifying these features helps locate genes within a genome. In bacteria, genes are often organized in operons with promoters and terminators, while eukaryotic genes have more complex regulatory elements and splicing signals.
Recommended video:
Guided course
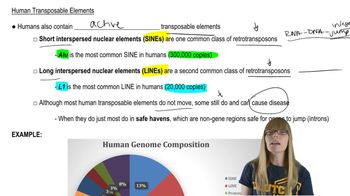
Human Transposable Elements
Differences Between Prokaryotic and Eukaryotic Genomes
Bacterial genomes are typically compact, with densely packed genes, few introns, and operon structures. Eukaryotic genomes are larger, contain introns, repetitive sequences, and complex regulatory regions. Recognizing these differences is essential for accurate gene annotation in each genome type.
Recommended video:
Guided course
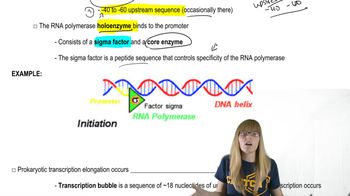
Prokaryotic Transcription
Sequence Features Indicative of Genes
Key sequence features include open reading frames (ORFs), start and stop codons, ribosome binding sites in bacteria, splice sites in eukaryotes, and conserved motifs. Detecting these features in unknown sequences aids in predicting gene locations and functions.
Recommended video:
Guided course

Sequencing Difficulties
Related Videos
Related Practice
Multiple Choice
499
views
1
rank


