The mature mRNA transcribed from the human β-globin gene is considerably longer than the sequence needed to encode the 146–amino acid polypeptide. Give the names of three sequences located on the mature β-globin mRNA but not translated.
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
11. Translation
The Genetic Code
Problem 29b
Textbook Question
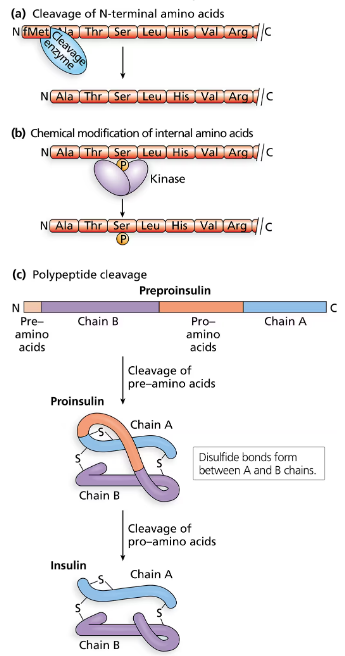
Explain why it is not feasible to insert the entire human insulin gene into E. coli and anticipate the production of insulin.
 Verified step by step guidance
Verified step by step guidance1
Understand that the human insulin gene contains both exons (coding regions) and introns (non-coding regions). In humans, the gene is transcribed into pre-mRNA, which then undergoes splicing to remove introns, producing mature mRNA.
Recognize that E. coli, being a prokaryote, lacks the cellular machinery to perform RNA splicing. Therefore, if the entire human insulin gene (including introns) is inserted into E. coli, the bacterium cannot correctly process the gene to produce functional mRNA.
Note that without proper mRNA processing, the translation machinery in E. coli will produce a nonfunctional or truncated protein because the introns will be translated as well, disrupting the insulin protein sequence.
Conclude that to produce insulin in E. coli, scientists use complementary DNA (cDNA) synthesized from mature mRNA, which contains only the exons, ensuring that the bacterial system can translate the gene correctly into functional insulin protein.
Summarize that the key issue is the presence of introns in the human insulin gene and the inability of E. coli to process these introns, making direct insertion of the entire gene ineffective for insulin production.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
1mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Gene Structure in Eukaryotes vs. Prokaryotes
Eukaryotic genes, like the human insulin gene, contain introns (non-coding regions) that are removed during RNA processing. Prokaryotes such as E. coli lack the machinery to splice out introns, so inserting the entire gene including introns prevents proper protein synthesis.
Recommended video:
Guided course

Chromosome Structure
Use of cDNA for Gene Expression in Bacteria
To express eukaryotic proteins in bacteria, scientists use complementary DNA (cDNA) synthesized from mature mRNA, which lacks introns. This allows E. coli to transcribe and translate the gene correctly, producing functional insulin.
Recommended video:
Guided course

Penetrance and Expressivity
Post-Translational Modifications and Protein Folding
Human insulin requires specific folding and modifications to become active, processes that E. coli may not perform correctly. Even with the gene expressed, bacterial systems might produce inactive or misfolded insulin without additional engineering.
Recommended video:
Guided course

Post Translational Modifications
Related Videos
Related Practice
Textbook Question
640
views


