In recombination studies of the rII locus in phage T4, what is the significance of the value determined by calculating phage growth in the K12 versus the B strains of E. coli following simultaneous infection in E. coli B? Which value is always greater?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
5. Genetics of Bacteria and Viruses
Bacteriophage Genetics
Problem 20a
Textbook Question
Using mutants 2 and 3 from Problem 19, following mixed infection on E. coli B, progeny viruses were plated in a series of dilutions on both E. coli B and K12 with the following results.
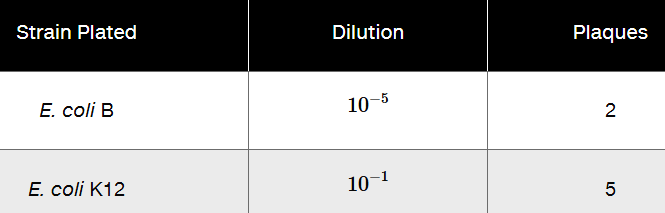
What is the recombination frequency between the two mutants?
 Verified step by step guidance
Verified step by step guidance1
Step 1: Understand the problem. The goal is to calculate the recombination frequency between two mutants based on the given plaque counts on two strains of E. coli (B and K12) at specific dilutions. Recombination frequency is calculated as the number of recombinant plaques divided by the total number of plaques, expressed as a percentage.
Step 2: Identify the total number of plaques. For E. coli B, the dilution is 10⁻⁵, and the number of plaques is 2. For E. coli K12, the dilution is 10⁻¹, and the number of plaques is 5. These values represent the observed plaques at the respective dilutions.
Step 3: Adjust the plaque counts to the same dilution factor. Since the dilutions are different (10⁻⁵ for E. coli B and 10⁻¹ for E. coli K12), scale the plaque counts to the same dilution factor. Use the formula: Adjusted Plaques = Observed Plaques × (Dilution Factor Adjustment).
Step 4: Determine the number of recombinant plaques. Recombinant plaques are those that grow on E. coli K12 but not on E. coli B. After adjusting the plaque counts to the same dilution factor, subtract the plaques observed on E. coli B from those observed on E. coli K12 to find the recombinant plaques.
Step 5: Calculate the recombination frequency. Use the formula: Recombination Frequency = (Number of Recombinant Plaques / Total Plaques) × 100. The total plaques are the sum of the adjusted plaques for E. coli B and E. coli K12. Express the result as a percentage.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
35sPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Recombination Frequency
Recombination frequency is a measure of the likelihood that two alleles will be separated during meiosis due to crossing over. It is calculated as the number of recombinant offspring divided by the total number of offspring, often expressed as a percentage. In the context of viral genetics, it helps determine the genetic distance between two mutants based on their progeny.
Recommended video:
Guided course

Recombination after Single Strand Breaks
Mutants in Genetics
Mutants are organisms that have undergone a change in their DNA sequence, resulting in a new phenotype. In the context of the question, mutants 2 and 3 refer to specific strains of viruses that have distinct genetic variations. Studying these mutants allows researchers to understand genetic interactions and the mechanisms of recombination.
Recommended video:
Guided course

The Genetic Code
Plaque Assay
A plaque assay is a method used to quantify the number of viral particles in a sample. It involves infecting a layer of host cells with diluted virus and observing the formation of plaques, which are clear zones indicating cell lysis due to viral infection. The number of plaques can be used to calculate viral titers and assess the effects of recombination between different viral mutants.
Recommended video:
Guided course

Plaques and Experiments

 3:44m
3:44mWatch next
Master Plaques and Experiments with a bite sized video explanation from Kylia
Start learningRelated Videos
Related Practice
Textbook Question
784
views
