Speculate on how improved living conditions and medical care in the developed nations might affect human mutation rates, both neutral and deleterious.
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
17. Mutation, Repair, and Recombination
Types of Mutations
Problem 22b
Textbook Question
The human β-globin wild-type allele and a certain mutant allele are identical in sequence except for a single base-pair substitution that changes one nucleotide at the end of intron 2. The wild-type and mutant sequences of the affected portion of pre-mRNA are
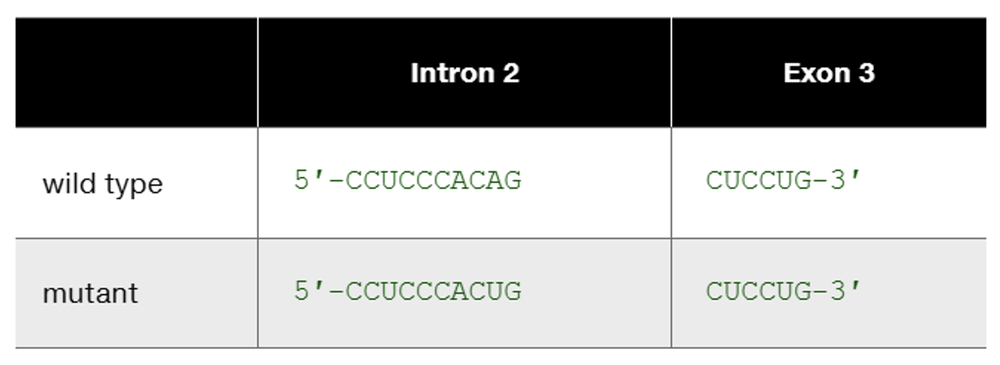
This is one example of how DNA sequence change occurring somewhere other than in an exon can produce mutation. List other kinds of DNA sequence changes occurring outside exons that can produce mutation. In each case, characterize the kind of change you would expect to see in mutant mRNA or mutant protein.
 Verified step by step guidance
Verified step by step guidance1
Understand the problem: The question asks for examples of DNA sequence changes outside exons that can lead to mutations, and how these changes affect mutant mRNA or protein. This involves understanding regulatory and structural elements of genes beyond coding regions.
Step 1: Splice site mutations - Explain that mutations in splice donor or acceptor sites (e.g., GT at the 5' end of an intron or AG at the 3' end) can disrupt splicing. This may result in intron retention, exon skipping, or the use of cryptic splice sites, leading to altered mRNA and potentially truncated or nonfunctional proteins.
Step 2: Promoter mutations - Discuss how mutations in the promoter region can affect transcription initiation. For example, a mutation in the TATA box or other regulatory elements can reduce or abolish transcription, leading to decreased or absent mRNA and protein production.
Step 3: Enhancer or silencer mutations - Highlight that mutations in enhancer or silencer regions can alter the binding of transcription factors, leading to changes in gene expression levels. This can result in overexpression, underexpression, or misexpression of the gene, affecting protein levels or function.
Step 4: Polyadenylation signal mutations - Explain that mutations in the polyadenylation signal (e.g., AAUAAA) can disrupt proper 3' end processing of mRNA. This can lead to unstable mRNA or altered protein products due to improper translation termination.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
1mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Introns and Exons
Introns are non-coding sequences in a gene that are transcribed into pre-mRNA but are removed during RNA splicing, while exons are the coding sequences that remain in the mature mRNA. Understanding the roles of introns and exons is crucial because mutations in introns can affect splicing, potentially leading to altered protein products even if the coding sequence remains unchanged.
Recommended video:
Guided course

mRNA Processing
Splice Site Mutations
Splice site mutations occur at the boundaries of introns and exons, affecting the recognition of these sites by the splicing machinery. Such mutations can lead to the inclusion of intronic sequences in the mRNA or the exclusion of exonic sequences, resulting in frameshift mutations or truncated proteins, which can have significant functional consequences.
Recommended video:
Guided course

Mutations and Phenotypes
Regulatory Element Mutations
Regulatory elements, such as enhancers and silencers, are DNA sequences that control the expression of genes. Mutations in these regions can alter the binding of transcription factors, leading to changes in gene expression levels. This can result in either overexpression or underexpression of the corresponding protein, potentially causing various phenotypic effects.
Recommended video:
Guided course
Human Transposable Elements
Related Videos
Related Practice
Textbook Question
535
views


