The line below represents a mature eukaryotic mRNA. The accompanying list contains many sequences or structures that are part of eukaryotic mRNA. A few of the items in the list, however, are not found in eukaryotic mRNA. As accurately as you can, show the location, on the line, of the sequences or structures that belong in eukaryotic mRNA; then, separately, list the items that are not part of eukaryotic mRNA.
5′ ____________________________ 3′
a. stop codon
b. poly-A tail
c. intron
d. 3' UTR
e. promoter
f. start codon
g. AAUAAA
h. 5' UTR
i. 5' cap
j. termination sequence
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
11. Translation
Translation
Problem 28
Textbook Question
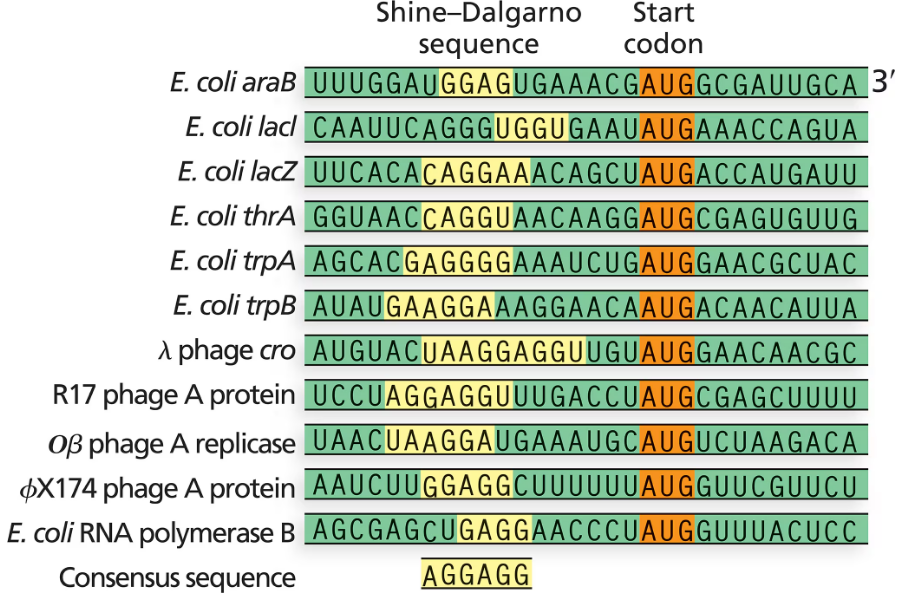
The following figure contains several examples of the Shine–Dalgarno sequence. Using the seven Shine–Dalgarno sequences from E. coli, determine the consensus sequence and describe its location relative to the start codon.
 Verified step by step guidance
Verified step by step guidance1
Step 1: Understand the Shine–Dalgarno sequence as a ribosomal binding site in prokaryotic mRNA, typically located upstream of the start codon, which helps initiate translation by aligning the ribosome with the start codon.
Step 2: Collect the seven Shine–Dalgarno sequences from the given E. coli examples and align them to identify conserved nucleotides across all sequences.
Step 3: For each position in the aligned sequences, determine the most frequently occurring nucleotide to establish the consensus sequence. This can be done by creating a position weight matrix or simply noting the dominant base at each position.
Step 4: Note the typical location of the Shine–Dalgarno sequence relative to the start codon, which is usually about 5 to 10 nucleotides upstream (5' direction) of the AUG start codon.
Step 5: Summarize the consensus sequence and describe its position relative to the start codon, emphasizing its role in facilitating ribosome binding and translation initiation.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
1mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Shine–Dalgarno Sequence
The Shine–Dalgarno sequence is a short, conserved ribosomal binding site in bacterial mRNA, typically located upstream of the start codon. It helps recruit the ribosome to initiate translation by base-pairing with a complementary sequence in the 16S rRNA of the small ribosomal subunit.
Recommended video:
Guided course

Sequencing Difficulties
Consensus Sequence
A consensus sequence represents the most common nucleotides found at each position in a set of similar sequences. It summarizes conserved features and is used to identify functional motifs, such as the Shine–Dalgarno sequence, by highlighting the typical nucleotide pattern.
Recommended video:
Guided course

Sequencing Overview
Relative Position to the Start Codon
The Shine–Dalgarno sequence is located a few nucleotides upstream (usually 5-10 bases) of the start codon (AUG). This precise positioning is critical for proper alignment of the ribosome with the start codon to ensure accurate initiation of protein synthesis.
Recommended video:
Guided course

Positional Cloning

 7:58m
7:58mWatch next
Master Translation initiation with a bite sized video explanation from Kylia
Start learningRelated Videos
Related Practice
Textbook Question
766
views
