Diagram a eukaryotic gene containing three exons and two introns, the pre-mRNA and mature mRNA transcript of the gene, and a partial polypeptide that contains the following sequences and features. Carefully align the nucleic acids, and locate each sequence or feature on the appropriate molecule.
a. The AG and GU dinucleotides corresponding to intron-exon junctions
b. The +1 nucleotide
c. The 5' UTR and the 3' UTR
d. The start codon sequence
e. A stop codon sequence
f. A codon sequence for the amino acids Gly-His-Arg at the end of exon 1 and a codon sequence for the amino acids Leu-Trp-Ala at the beginning of exon 2
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
11. Translation
The Genetic Code
Problem 41d
Textbook Question
The two gels illustrated contain dideoxynucleotide DNA-sequencing information for a wild-type segment and mutant segment of DNA corresponding to the N-terminal end of a protein. The start codon and the next five codons are sequenced.
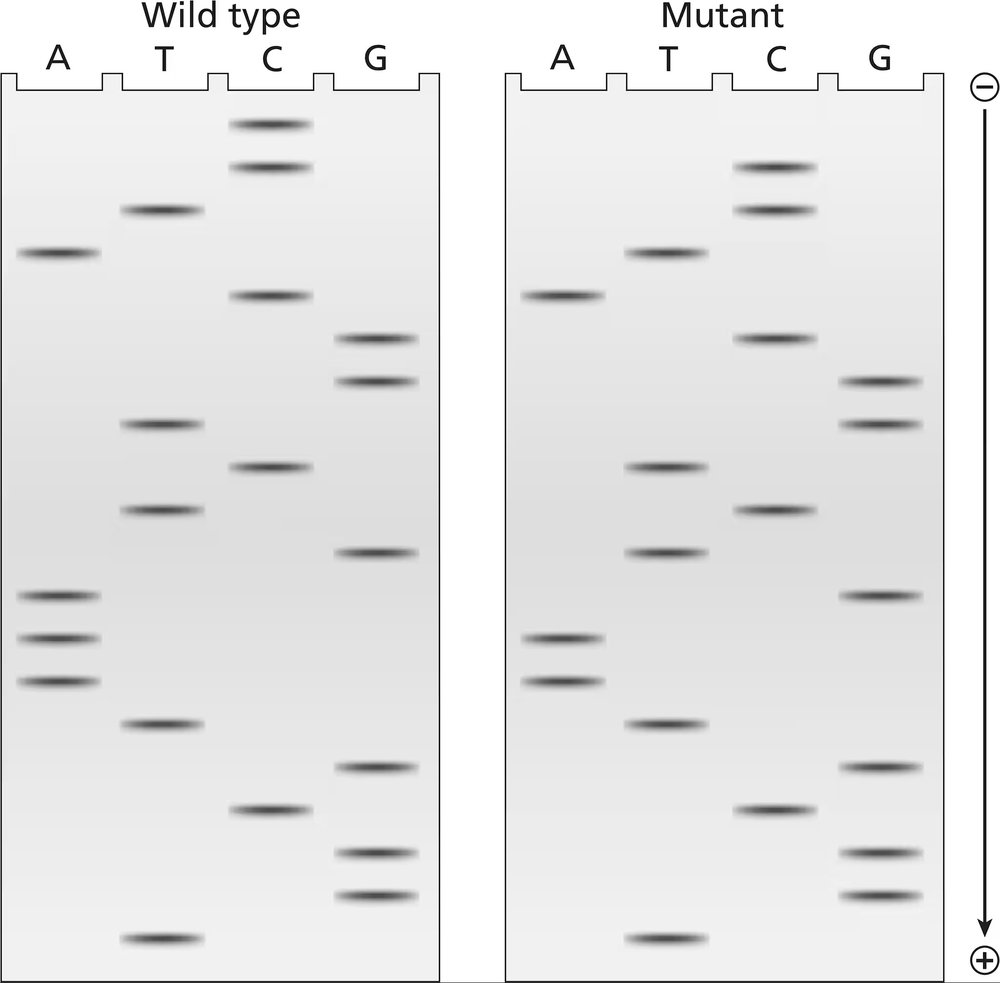
Determine the amino acid sequences translated from these mRNAs.
 Verified step by step guidance
Verified step by step guidance1
Examine the DNA sequencing gels for both the wild-type and mutant segments. Identify the sequence of nucleotides (A, T, G, C) for each strand by reading the gel from bottom to top, as smaller fragments migrate further in the gel.
Convert the DNA sequences obtained from the gels into their corresponding mRNA sequences. Replace thymine (T) in the DNA sequence with uracil (U) to form the mRNA sequence.
Identify the start codon (AUG) in the mRNA sequence, as this marks the beginning of translation. Divide the mRNA sequence into codons (groups of three nucleotides) starting from the AUG codon.
Using the genetic code table, translate each codon into its corresponding amino acid. Continue translating until you encounter a stop codon (UAA, UAG, or UGA), which signals the end of translation.
Compare the amino acid sequences derived from the wild-type and mutant mRNA sequences. Note any differences in the amino acid sequence caused by the mutation, and consider how these changes might affect the protein's function.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
1mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Dideoxynucleotide Sequencing
Dideoxynucleotide sequencing, also known as Sanger sequencing, is a method used to determine the nucleotide sequence of DNA. It involves incorporating dideoxynucleotides, which terminate DNA strand elongation, allowing for the generation of fragments of varying lengths. By analyzing these fragments through gel electrophoresis, researchers can deduce the sequence of the original DNA template.
Recommended video:
Guided course

Sanger Sequencing
Codons and Translation
Codons are sequences of three nucleotides in mRNA that correspond to specific amino acids during protein synthesis. The process of translation occurs in ribosomes, where the mRNA is read in sets of codons, and transfer RNA (tRNA) molecules bring the appropriate amino acids to form a polypeptide chain. Understanding codons is essential for translating mRNA into the correct amino acid sequence.
Recommended video:
Guided course

Translation initiation
Wild-Type vs. Mutant DNA
Wild-type DNA refers to the normal, non-mutated version of a gene, while mutant DNA contains alterations that may affect gene function. These mutations can lead to changes in the amino acid sequence of proteins, potentially impacting their structure and function. Analyzing both wild-type and mutant sequences is crucial for understanding the effects of genetic variations on protein translation.
Recommended video:
Guided course

Mutations and Phenotypes
Related Videos
Related Practice
Textbook Question
884
views


