Define antisense RNA, and describe how it affects the translation of a complementary mRNA. Why is it more advantageous to the organism to stop translation initiation than to inactivate or destroy the gene product after it is produced?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
12. Gene Regulation in Prokaryotes
Riboswitches
Problem 29a
Textbook Question
The bacterial insertion sequence IS10 uses antisense RNA to regulate translation of the mRNA that produces the enzyme transposase, which is required for insertion sequence transposition. Transcription of the antisense RNA gene is controlled by POUT, which is more than 10 times more efficient at transcription than the PIN promoter, which controls transposase gene transcription.
If a mutation reduced the transcriptional efficiency of POUT so as to be equal to that of PIN, what is the likely effect on the transposition of IS10?
 Verified step by step guidance
Verified step by step guidance1
Understand the role of antisense RNA in regulating transposase production. Antisense RNA binds to the mRNA of the transposase gene, preventing its translation into the transposase enzyme. This regulation ensures controlled transposition activity of IS10.
Recognize the significance of the POUT promoter. POUT drives the transcription of the antisense RNA gene and is normally much more efficient than the PIN promoter, which drives the transcription of the transposase gene. This imbalance ensures that antisense RNA is produced in higher quantities, effectively suppressing transposase production.
Analyze the mutation described in the problem. The mutation reduces the transcriptional efficiency of POUT, making it equal to that of PIN. This means that the production of antisense RNA will decrease, while the production of transposase mRNA remains unchanged.
Predict the effect of reduced antisense RNA levels. With less antisense RNA available to bind to transposase mRNA, more transposase enzyme will be produced. This increase in transposase levels will likely lead to higher rates of IS10 transposition.
Conclude the likely outcome. The mutation disrupts the regulatory balance between antisense RNA and transposase mRNA, leading to increased transposition activity of IS10, which could have significant effects on the bacterial genome, such as increased genetic instability or altered gene expression.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
4mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Antisense RNA
Antisense RNA is a strand of RNA that is complementary to a specific mRNA strand. It can bind to the mRNA and inhibit its translation, effectively regulating gene expression. In the context of IS10, the antisense RNA regulates the translation of transposase, which is crucial for the transposition process.
Recommended video:
Transcriptional Efficiency
Transcriptional efficiency refers to the rate at which a gene is transcribed into mRNA. It is influenced by various factors, including promoter strength and regulatory elements. In this scenario, the POUT promoter is significantly more efficient than the PIN promoter, affecting the levels of antisense RNA and transposase produced, which in turn impacts transposition.
Recommended video:
Guided course

Eukaryotic Transcription
Transposition
Transposition is the process by which a transposable element, such as IS10, moves from one location in the genome to another. This process is mediated by the enzyme transposase, which is produced in response to the transcription of the transposase gene. The efficiency of transposition is directly linked to the levels of transposase and the regulatory mechanisms controlling its expression.
Recommended video:
Guided course
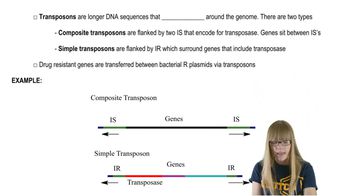
Prokaryotic Transposable Elements
Related Videos
Related Practice
Textbook Question
1058
views



