Contrast the structure of SINE and LINE DNA sequences. Why are LINEs referred to as retrotransposons?

How many base pairs are in a molecule of phage T2 DNA 52-µm long?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
DNA Structure and Base Pairing

DNA Length Measurement and Conversion
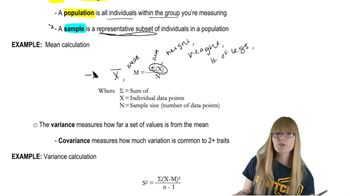
Phage T2 DNA Characteristics

Mammals contain a diploid genome consisting of at least 10⁹ bp. If this amount of DNA is present as chromatin fibers, where each group of 200 bp of DNA is combined with 9 histones into a nucleosome and each group of 6 nucleosomes is combined into a solenoid, achieving a final packing ratio of 50, determine:
(a) the total number of nucleosomes in all fibers,
(b) the total number of histone molecules combined with DNA in the diploid genome, and
(c) the combined length of all fibers.
Assume that a viral DNA molecule is a 50-µm-long circular strand with a uniform 20-Å diameter. If this molecule is contained in a viral head that is a 0.08-µm-diameter sphere, will the DNA molecule fit into the viral head, assuming complete flexibility of the molecule? Justify your answer mathematically.
The human genome contains approximately 106 copies of an Alu sequence, one of the best-studied classes of short interspersed elements (SINEs), per haploid genome. Individual Alu units share a 282-nucleotide consensus sequence followed by a 3'-adenine-rich tail region [Schmid (1998)]. Given that there are approximately 3 x 109 base pairs per human haploid genome, about how many base pairs are spaced between each Alu sequence?
The following is a diagram of the general structure of the bacteriophage chromosome. Speculate on the mechanism by which it forms a closed ring upon infection of the host cell.
