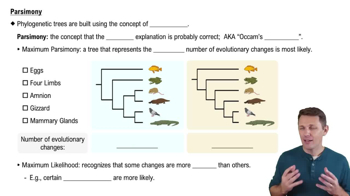
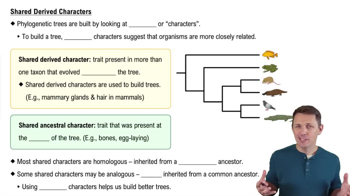
In a comparison of birds and mammals, the condition of having four limbs is
a. A shared ancestral character
b. A shared derived character
c. A character useful for distinguishing birds from mammals
d. An example of analogy rather than homology

 Verified step by step guidance
Verified step by step guidance

In a comparison of birds and mammals, the condition of having four limbs is
a. A shared ancestral character
b. A shared derived character
c. A character useful for distinguishing birds from mammals
d. An example of analogy rather than homology
Which similarly inclusive taxon is represented as descending from the same common ancestor as Canidae?
<IMAGE>
a. Felidae
b. Mustelidae
c. Carnivora
d. Lutra
Three living species, X, Y, and Z, share a common ancestor, T, as do extinct species U and V. A grouping that consists of species T, X, Y, and Z (but not U or V) makes up
a. A monophyletic taxon
b. An ingroup, with species U as the outgroup
c. A paraphyletic group
d. A polyphyletic group
Based on the tree below, which statement is not correct? <IMAGE>
a. Goats and humans form a sister group
b. Salamanders are a sister group to the group containing lizards, goats, and humans
c. Salamanders are as closely related to goats as to humans
d. Lizards are more closely related to salamanders than to humans