Should the FDA regulate direct-to-consumer genetic tests, or should these tests be available as a 'buyer beware' product?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
18. Molecular Genetic Tools
Methods for Analyzing DNA
Problem 21b
Textbook Question
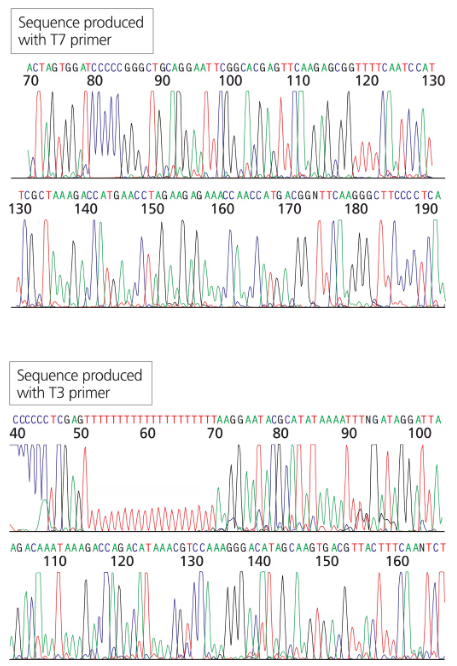
You have isolated another cDNA clone of the CRABS CLAW gene from a cDNA library. The cDNA was directionally cloned using the EcoRI and XhoI sites. You sequence the recombinant plasmid using primers complementary to the T7 and T3 promoter sites flanking the MCS. The first 30 to 60 bases of sequence are usually discarded since they tend to contain errors.
Will the long stretch of T residues in the T3 sequence exist in the genomic sequence of the gene?
 Verified step by step guidance
Verified step by step guidance1
Understand the context: The cDNA clone of the CRABS CLAW gene was constructed using a cDNA library. cDNA is synthesized from mRNA, meaning it represents the expressed sequences of a gene (exons only) and does not include introns or regulatory sequences found in the genomic DNA.
Analyze the T3 promoter sequence: The T3 promoter is part of the vector used for cloning and sequencing. It is not derived from the genomic DNA or the cDNA of the gene itself. Instead, it is an artificial sequence included in the vector to facilitate transcription in vitro.
Determine the origin of the long stretch of T residues: The long stretch of T residues in the T3 promoter sequence is part of the vector design and does not originate from the genomic sequence of the CRABS CLAW gene. It is an engineered sequence used for experimental purposes.
Compare genomic DNA and cDNA: Genomic DNA contains both exons and introns, as well as regulatory regions. cDNA, on the other hand, is synthesized from mRNA and represents only the coding regions (exons). The T3 promoter sequence is unrelated to either genomic DNA or cDNA, as it is part of the vector.
Conclude: The long stretch of T residues in the T3 promoter sequence will not exist in the genomic sequence of the CRABS CLAW gene because it is an artificial sequence included in the vector and not part of the gene's natural genomic or cDNA sequence.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
2mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
cDNA and Genomic DNA
cDNA (complementary DNA) is synthesized from mRNA through reverse transcription and represents only the expressed genes, lacking introns. In contrast, genomic DNA contains both coding (exons) and non-coding regions (introns) of the entire genome. Understanding the difference is crucial for determining whether sequences found in cDNA, like the T residues in the T3 promoter, will be present in the genomic DNA.
Recommended video:
Guided course

Functional Genomics
Promoter Sequences
Promoter sequences are regions of DNA that initiate transcription of a gene. They are typically located upstream of the coding region and are recognized by RNA polymerase and transcription factors. The T3 promoter, which contains a long stretch of T residues, is used in cloning and expression systems, but its presence in genomic DNA depends on the specific gene structure and regulatory elements.
Recommended video:
Guided course

Sequencing Difficulties
Sequencing and Error Correction
Sequencing involves determining the order of nucleotides in DNA. In this context, the first 30 to 60 bases of sequence are often discarded due to potential errors, which can arise from the sequencing process itself. This practice is important for ensuring the accuracy of the data, especially when analyzing sequences that may include repetitive elements like long stretches of T residues, which can complicate interpretation.
Recommended video:
Guided course

Sequencing Difficulties

 7:40m
7:40mWatch next
Master Methods for Analyzing DNA and RNA with a bite sized video explanation from Kylia
Start learningRelated Videos
Related Practice
Textbook Question
507
views
