What are the two groups of reproductive isolating mechanisms? Which of these is regarded as more efficient, and why?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
21. Population Genetics
Allelic Frequency Changes
Problem 33b
Textbook Question
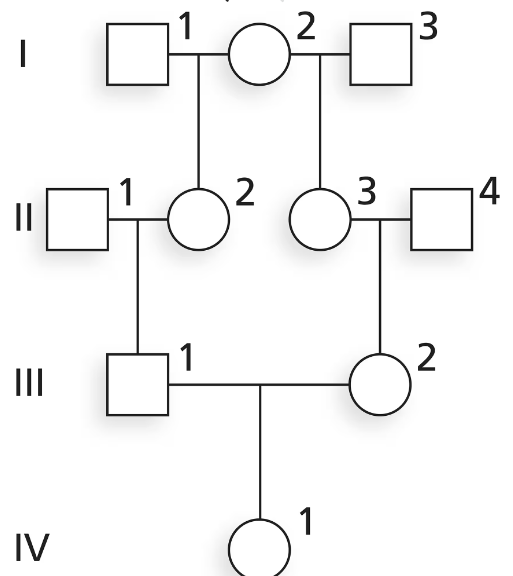
Evaluate the following pedigree, and answer the questions below for individual IV-1. What is F for this individual?
 Verified step by step guidance
Verified step by step guidance1
Step 1: Understand the concept of 'F', which represents the inbreeding coefficient. It measures the probability that an individual has two alleles at a locus that are identical by descent due to shared ancestry.
Step 2: Analyze the pedigree provided. Identify all ancestors of individual IV-1 and determine if there are any loops in the pedigree (i.e., instances where the same ancestor appears more than once due to consanguineous mating).
Step 3: For each loop in the pedigree, calculate the contribution to the inbreeding coefficient using the formula: , where n is the number of individuals in the loop excluding the individual being evaluated.
Step 4: Sum the contributions from all loops to determine the total inbreeding coefficient for individual IV-1. Ensure that you account for all possible paths of shared ancestry.
Step 5: Verify your calculations by cross-checking the pedigree and ensuring that all loops and shared ancestors have been accounted for correctly.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
1mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Pedigree Analysis
Pedigree analysis is a method used to study the inheritance patterns of traits in families. It involves creating a diagram that represents family relationships and the presence or absence of specific traits across generations. This visual representation helps in understanding how traits are passed down and can indicate whether a trait is dominant, recessive, or linked to sex chromosomes.
Recommended video:
Guided course

Pedigree Flowchart
Coefficient of Inbreeding (F)
The coefficient of inbreeding (F) quantifies the probability that an individual has inherited identical alleles from both parents due to a common ancestor. It ranges from 0 (no inbreeding) to 1 (complete inbreeding). In pedigree analysis, calculating F helps assess the genetic diversity of a population and the potential for genetic disorders resulting from inbreeding.
Recommended video:
Guided course

F Factor and Hfr
Generational Relationships
Generational relationships in a pedigree chart indicate the lineage and connections between individuals. Each generation is typically represented by a horizontal row, with parents above their offspring. Understanding these relationships is crucial for determining the inheritance patterns of traits and calculating coefficients like F, as it allows for the identification of common ancestors and the degree of relatedness between individuals.
Recommended video:
Guided course
Phylogenetic Trees
Related Videos
Related Practice
Textbook Question
594
views


