Present an overview of various forms of posttranscriptional RNA processing in eukaryotes. For each, provide an example.
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
10. Transcription
RNA Modification and Processing
Problem 30e
Textbook Question
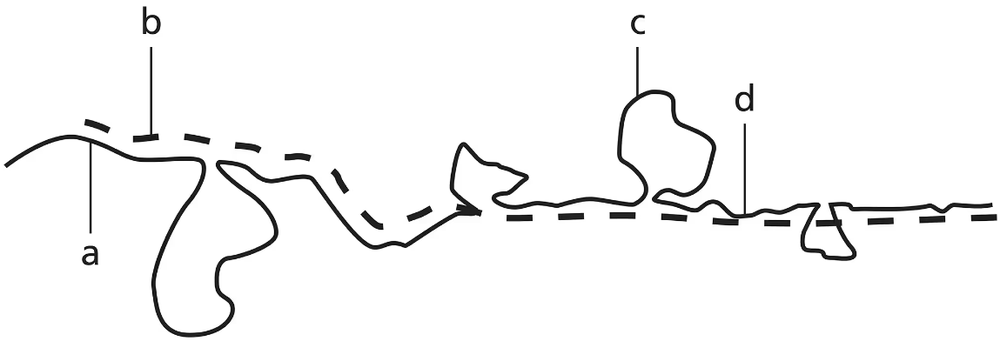
Genomic DNA from a mouse is isolated, fragmented, and denatured into single strands. It is then mixed with mRNA isolated from the cytoplasm of mouse cells. The image represents an electron micrograph result showing the hybridization of single-stranded DNA and mRNA. Based on this electron micrograph image, how many introns and exons are present in the mouse DNA fragment shown?
 Verified step by step guidance
Verified step by step guidance1
Understand the context: The problem involves analyzing an electron micrograph showing hybridization between single-stranded DNA and mRNA. This technique is used to identify the regions of DNA that correspond to exons (coding regions) and introns (non-coding regions).
Recall the principle of hybridization: mRNA hybridizes only to the complementary exonic regions of the DNA, as introns are spliced out during mRNA processing. Introns will appear as loops in the DNA strand because they do not have complementary sequences in the mRNA.
Examine the electron micrograph: Count the number of loops (representing introns) and the number of hybridized regions (representing exons). Each loop corresponds to an intron, and each hybridized segment corresponds to an exon.
Determine the total number of introns and exons: The total number of exons is equal to the number of hybridized regions, and the total number of introns is equal to the number of loops observed in the micrograph.
Summarize the findings: Based on the counts from the micrograph, you can determine the number of introns and exons in the DNA fragment. Ensure that you clearly distinguish between the two types of regions in your analysis.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
3mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Introns and Exons
Introns and exons are segments of a gene found in eukaryotic DNA. Exons are the coding regions that are expressed and translated into proteins, while introns are non-coding regions that are removed during RNA processing. Understanding the distinction between these two is crucial for analyzing gene structure and function.
Recommended video:
Guided course

mRNA Processing
Hybridization
Hybridization refers to the process where complementary strands of nucleic acids, such as DNA and RNA, bind together. In the context of the question, the hybridization of single-stranded DNA with mRNA indicates the regions of the DNA that correspond to expressed genes, which can help identify exons and infer the presence of introns.
Recommended video:
Guided course

Drosophila P Element
Electron Micrography
Electron micrography is a technique that uses electron beams to visualize samples at a very high resolution. In this context, it allows researchers to observe the hybridized DNA and mRNA, providing insights into the structure of the genetic material and the arrangement of introns and exons within the DNA fragment.
Recommended video:
Guided course
Induced Mutations
Related Videos
Related Practice
Textbook Question
452
views


