Genomic DNA from a mouse is isolated, fragmented, and denatured into single strands. It is then mixed with mRNA isolated from the cytoplasm of mouse cells. The image represents an electron micrograph result showing the hybridization of single-stranded DNA and mRNA. Based on this electron micrograph image, how many introns and exons are present in the mouse DNA fragment shown?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
10. Transcription
RNA Modification and Processing
Problem 33b
Textbook Question
Isoginkgetin is a cell-permeable chemical isolated from the Ginkgo biloba tree that binds to and inhibits snRNPs.
Would this be most problematic for E. coli cells, yeast cells, or human cells? Why?
 Verified step by step guidance
Verified step by step guidance1
Understand the role of snRNPs (small nuclear ribonucleoproteins): These are essential components of the spliceosome, which is responsible for the removal of introns from pre-mRNA during RNA splicing in eukaryotic cells.
Recognize that E. coli cells are prokaryotic and do not have introns in their genes. Therefore, they do not require snRNPs or a spliceosome for gene expression.
Identify that yeast cells are eukaryotic and, like human cells, have introns in their genes. They rely on snRNPs for RNA splicing to produce mature mRNA.
Understand that human cells, being eukaryotic, also depend on snRNPs for RNA splicing. However, human cells typically have more complex splicing requirements due to the presence of alternative splicing, which allows a single gene to produce multiple proteins.
Conclude that the inhibition of snRNPs by isoginkgetin would not affect E. coli cells but would be problematic for yeast and human cells. Among these, human cells might be more severely affected due to their reliance on alternative splicing for generating protein diversity.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
3mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
snRNPs (small nuclear ribonucleoproteins)
snRNPs are essential components of the spliceosome, a complex responsible for the splicing of pre-mRNA in eukaryotic cells. They play a critical role in the removal of introns and the joining of exons, which is vital for producing mature mRNA that can be translated into proteins.
Recommended video:
Guided course

mRNA Processing
Eukaryotic vs. Prokaryotic Cells
Eukaryotic cells, such as those in yeast and humans, have a defined nucleus and complex organelles, while prokaryotic cells, like E. coli, lack a nucleus and have a simpler structure. This distinction is crucial because snRNPs are primarily involved in processes that occur in eukaryotic cells, making them less relevant in prokaryotic organisms.
Recommended video:
Guided course
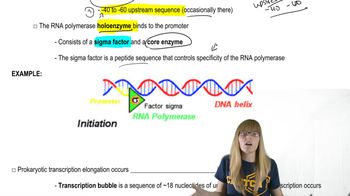
Prokaryotic Transcription
Impact of Inhibiting snRNPs
Inhibiting snRNPs would disrupt the splicing process in eukaryotic cells, leading to the production of improperly processed mRNA and potentially nonfunctional proteins. This disruption can severely affect cellular function and viability, particularly in human and yeast cells, which rely on accurate mRNA splicing for gene expression.
Recommended video:
Guided course

Segmentation Genes
Related Videos
Related Practice
Textbook Question
725
views


