An article entitled 'Nucleosome Positioning at the Replication Fork' states: 'both the 'old' randomly segregated nucleosomes as well as the 'new' assembled histone octamers rapidly position themselves (within seconds) on the newly replicated DNA strands' [Lucchini et al. (2002)]. Given this statement, how would one compare the distribution of nucleosomes and DNA in newly replicated chromatin? How could one experimentally test the distribution of nucleosomes on newly replicated chromosomes?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
7. DNA and Chromosome Structure
Eukaryotic Chromosome Structure
Problem 27
Textbook Question
Spermatogenesis in mammals results in sperm that have a nucleus that is 40 times smaller than an average somatic cell. Thus, the sperm haploid genome must be packaged very tightly, yet in a way that is reversible after fertilization. This sperm-specific DNA compaction is due to a nucleosome-to-nucleoprotamine transition, where the histone-based nucleosomes are removed and replaced with arginine-rich protamine proteins that facilitate a tighter packaging of DNA. In 2013 Montellier et al. showed that replacement of the H2B protein in the nucleosomes with a testis-specific variant of H2B called TSH2B is a critical step prior to the nucleosome-to-nucleoprotamine transition. Mice lacking TSH2B retain H2B and their sperm arrest late in spermatogenesis with reduced DNA compaction. Based on these findings, would you expect that TSH2B-containing nucleosomes are more or less stable than H2B-containing nucleosomes? Explain your reasoning.
 Verified step by step guidance
Verified step by step guidance1
Step 1: Begin by understanding the role of TSH2B in the nucleosome-to-nucleoprotamine transition. TSH2B is a testis-specific variant of the H2B protein that replaces H2B in nucleosomes during spermatogenesis. This replacement is critical for facilitating the transition to protamine-based DNA packaging.
Step 2: Consider the findings from Montellier et al. (2013). Mice lacking TSH2B retain H2B in their nucleosomes, and their sperm exhibit reduced DNA compaction and arrest late in spermatogenesis. This suggests that TSH2B plays a key role in enabling tighter DNA packaging.
Step 3: Analyze the stability of TSH2B-containing nucleosomes compared to H2B-containing nucleosomes. Since TSH2B facilitates the removal of histone-based nucleosomes and the transition to protamine-based packaging, it is likely that TSH2B-containing nucleosomes are less stable than H2B-containing nucleosomes. This reduced stability may make it easier for the nucleosomes to be disassembled during the transition.
Step 4: Consider the biochemical properties of TSH2B. Testis-specific histone variants like TSH2B are often adapted to the unique requirements of spermatogenesis, including facilitating DNA compaction and transitions. The reduced stability of TSH2B-containing nucleosomes aligns with the need for reversible DNA packaging during fertilization.
Step 5: Conclude that the reduced stability of TSH2B-containing nucleosomes is a functional adaptation that supports the nucleosome-to-nucleoprotamine transition. This allows for the tight yet reversible DNA packaging required in sperm cells, ensuring proper function during fertilization.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
1mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Spermatogenesis
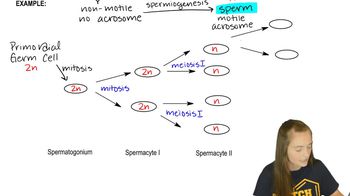
Spermatogenesis is the process by which male gametes, or sperm cells, are produced in the testes. It involves several stages, including mitosis, meiosis, and spermiogenesis, leading to the formation of mature sperm. This process is crucial for sexual reproduction and is characterized by significant cellular changes, including DNA compaction to fit within the small sperm nucleus.
Recommended video:
Guided course
Gamete Development
Nucleosome-to-Nucleoprotamine Transition
The nucleosome-to-nucleoprotamine transition is a critical step in sperm development where histone proteins in nucleosomes are replaced by protamines. This transition allows for a much tighter packaging of DNA, which is essential for the small size of the sperm nucleus. Protamines, being arginine-rich, facilitate this compaction, making the DNA more stable and less accessible, which is important for protecting the genetic material during transit.
Recommended video:
Guided course

Chromatin
Histone Variants and Stability
Histone variants, such as TSH2B, can influence the stability and function of nucleosomes. The presence of specific variants can alter the interactions between DNA and histones, potentially affecting the overall stability of the nucleosome structure. In the context of spermatogenesis, the replacement of H2B with TSH2B may enhance the stability of nucleosomes, thereby facilitating the transition to a more compact form necessary for sperm development.
Recommended video:
Guided course

Histone Protein Modifications
Related Videos
Related Practice
Textbook Question
492
views


