Private companies are offering personal DNA sequencing along with interpretation. What services do they offer? Do you think that these services should be regulated, and if so, in what way? Investigate one such company, 23andMe, at http://www.23andMe.com, before answering these questions.
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
18. Molecular Genetic Tools
Methods for Analyzing DNA
Problem 27a
Textbook Question
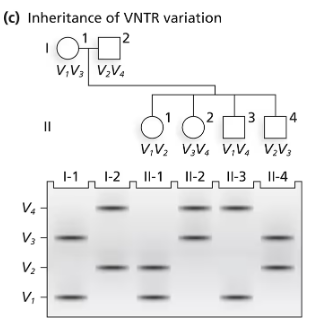
A family consisting of a mother (I-1), a father (I-2), and three children (II-1, II-2, and II-3) are genotyped by PCR for a region of an autosome containing repeats of a 10-bp sequence. The mother carries 16 repeats on one chromosome and 21 on the homologous chromosome. The father carries repeat numbers of 18 and 26.
Following the layout of the following figure, which aligns members of a pedigree with their DNA fragments in a gel, draw a DNA gel containing the PCR fragments generated by amplification of DNA from the parents (I-1 and I-2). Label the size of each fragment.
 Verified step by step guidance
Verified step by step guidance1
Understand the problem: The goal is to create a DNA gel representation of PCR fragments for the parents (I-1 and I-2) based on the number of repeats in a specific region of their DNA. Each repeat is 10 base pairs (bp) long, and the total fragment size will depend on the number of repeats plus the flanking regions amplified by PCR.
Calculate the fragment sizes for the mother (I-1): The mother has 16 repeats on one chromosome and 21 repeats on the homologous chromosome. Multiply the number of repeats by 10 bp/repeat to determine the size contributed by the repeats. Add the size of the flanking regions (constant for all fragments, typically provided in the problem or assumed to be a standard value). Represent these two fragment sizes on the gel.
Calculate the fragment sizes for the father (I-2): The father has 18 repeats on one chromosome and 26 repeats on the homologous chromosome. Similarly, multiply the number of repeats by 10 bp/repeat and add the size of the flanking regions. Represent these two fragment sizes on the gel.
Draw the gel layout: On the gel, each lane corresponds to an individual. For the parents, label the lanes as I-1 (mother) and I-2 (father). Place the calculated fragment sizes in their respective lanes, with smaller fragments migrating farther down the gel and larger fragments staying closer to the top. Label the size of each fragment next to the bands.
Ensure proper labeling and alignment: Clearly label the lanes with the individual identifiers (I-1 and I-2) and the fragment sizes. Align the bands vertically to indicate the relative sizes of the fragments. This will allow for easy comparison of the DNA fragments between the parents.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
2mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Polymerase Chain Reaction (PCR)
PCR is a molecular biology technique used to amplify specific DNA sequences, making millions of copies of a particular segment. It involves repeated cycles of denaturation, annealing, and extension, allowing researchers to analyze genetic material in detail. Understanding PCR is crucial for interpreting the results of genetic tests, such as those used in this pedigree analysis.
Recommended video:
Guided course
Genetic Cloning
DNA Fragment Size and Gel Electrophoresis
Gel electrophoresis is a method used to separate DNA fragments based on their size. When DNA is subjected to an electric field in a gel matrix, smaller fragments move faster than larger ones, allowing for size estimation. This technique is essential for visualizing PCR products and determining the number of repeats in the DNA sequences from the parents.
Recommended video:
Guided course
Methods for Analyzing DNA and RNA
Genetic Inheritance Patterns
Genetic inheritance patterns describe how traits and genetic information are passed from parents to offspring. In this case, the number of repeats in the DNA sequences can exhibit co-dominance, where both parental alleles contribute to the phenotype. Understanding these patterns is vital for predicting the potential genotypes of the children based on the parents' genotypes.
Recommended video:
Guided course

Diploid Genetics

 7:40m
7:40mWatch next
Master Methods for Analyzing DNA and RNA with a bite sized video explanation from Kylia
Start learningRelated Videos
Related Practice
Textbook Question
503
views
