The ability to taste the bitter compound phenylthiocarbamide (PTC) is an autosomal dominant trait. The inability to taste PTC is a recessive condition. In a sample of 500 people, 360 have the ability to taste PTC and 140 do not. Calculate the frequency of each genotype.
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
21. Population Genetics
Hardy Weinberg
Problem 10a
Textbook Question
Consider a population in which the frequency of allele A is p=0.7 and the frequency of allele a is q=0.3 and where the alleles are codominant. What will be the allele frequencies after one generation if the following occurs?
wAA=1, wAa=0.9, waa=0.8
 Verified step by step guidance
Verified step by step guidance1
Identify the initial allele frequencies: p = 0.7 for allele A and q = 0.3 for allele a. Since p + q = 1, these represent the starting frequencies before selection.
Calculate the genotype frequencies under Hardy-Weinberg equilibrium before selection: . Substitute the values of p and q to get these frequencies.
Apply the fitness values to each genotype to find the weighted genotype frequencies after selection: multiply each genotype frequency by its respective fitness (wAA = 1, wAa = 0.9, waa = 0.8). This gives the adjusted frequencies reflecting survival or reproductive success.
Calculate the mean fitness of the population, denoted as , by summing the weighted genotype frequencies. This value normalizes the frequencies to ensure they sum to 1 after selection.
Determine the new allele frequencies after selection by calculating the proportion of each allele in the surviving population: and . These formulas account for the contribution of heterozygotes to each allele frequency.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
57sPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Allele Frequencies and Hardy-Weinberg Principle
Allele frequencies represent the proportion of different alleles in a population. The Hardy-Weinberg principle predicts genotype frequencies from allele frequencies under no evolutionary forces. Understanding initial allele frequencies (p and q) is essential to calculate expected genotype distributions before selection.
Recommended video:
Guided course

Hardy Weinberg
Codominance
Codominance occurs when both alleles in a heterozygote are fully expressed, producing a distinct phenotype for heterozygotes. This affects how genotypes contribute to the population's traits and fitness, influencing how selection acts on each genotype.
Recommended video:
Guided course
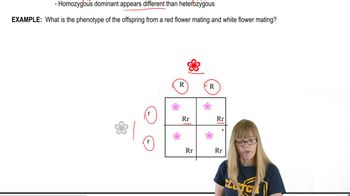
Variations on Dominance
Selection Coefficients and Fitness
Fitness values (w) measure the reproductive success of genotypes. Selection changes allele frequencies by favoring genotypes with higher fitness. Calculating post-selection allele frequencies requires weighting genotype frequencies by their fitness and normalizing to find new allele proportions.
Recommended video:
Guided course
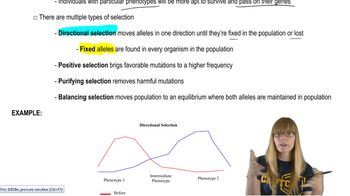
Natural Selection
Related Videos
Related Practice
Textbook Question
1065
views


