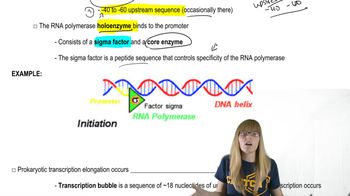
Describe the structure of RNA polymerase in bacteria. What is the core enzyme? What is the role of the σ subunit?

One form of posttranscriptional modification of most eukaryotic pre-mRNAs is the addition of a poly-A sequence at the 3' end. The absence of a poly-A sequence leads to rapid degradation of the transcript. Poly-A sequences of various lengths are also added to many bacterial RNA transcripts where, instead of promoting stability, they enhance degradation. In both cases, RNA secondary structures, stabilizing proteins, or degrading enzymes interact with poly-A sequences. Considering the activities of RNAs, what might be general functions of 3'-polyadenylation?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Posttranscriptional Modification and Polyadenylation

Differences in Polyadenylation between Eukaryotes and Prokaryotes
Role of RNA Secondary Structures and Protein Interactions

Write a paragraph describing the abbreviated chemical reactions that summarize RNA polymerase-directed transcription.
Messenger RNA molecules are very difficult to isolate in bacteria because they are rather quickly degraded in the cell. Can you suggest a reason why this occurs? Eukaryotic mRNAs are more stable and exist longer in the cell than do bacterial mRNAs. Is this an advantage or a disadvantage for a pancreatic cell making large quantities of insulin?
In a mixed copolymer experiment, messages were created with either 4/5C:1/5A or 4/5A:1/5C. These messages yielded proteins with the following amino acid compositions.
Using these data, predict the most specific coding composition for each amino acid.
Shown here are the amino acid sequences of the wild-type and three mutant forms of a short protein.
___________________________________________________
Wild-type: Met-Trp-Tyr-Arg-Gly-Ser-Pro-Thr
Mutant 1: Met-Trp
Mutant 2: Met-Trp-His-Arg-Gly-Ser-Pro-Thr
Mutant 3: Met-Cys-Ile-Val-Val-Val-Gln-His _
Use this information to answer the following questions:
Using the genetic coding dictionary, predict the type of mutation that led to each altered protein.
Shown here are the amino acid sequences of the wild-type and three mutant forms of a short protein.
___________________________________________________
Wild-type: Met-Trp-Tyr-Arg-Gly-Ser-Pro-Thr
Mutant 1: Met-Trp
Mutant 2: Met-Trp-His-Arg-Gly-Ser-Pro-Thr
Mutant 3: Met-Cys-Ile-Val-Val-Val-Gln-Hi
___________________________________________________
Use this information to answer the following questions:
For each mutant protein, determine the specific ribonucleotide change that led to its synthesis.
