
The human genome contains approximately 106 copies of an Alu sequence, one of the best-studied classes of short interspersed elements (SINEs), per haploid genome. Individual Alu units share a 282-nucleotide consensus sequence followed by a 3'-adenine-rich tail region [Schmid (1998)]. Given that there are approximately 3 x 109 base pairs per human haploid genome, about how many base pairs are spaced between each Alu sequence?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Human Genome Size and Structure

Alu Sequences and Short Interspersed Elements (SINEs)
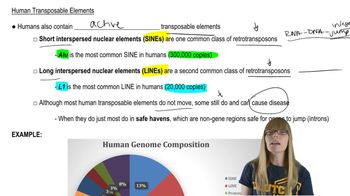
Calculating Average Spacing Between Repetitive Elements
While much remains to be learned about the role of nucleosomes and chromatin structure and function, recent research indicates that in vivo chemical modification of histones is associated with changes in gene activity. One study determined that acetylation of H3 and H4 is associated with 21.1 percent and 13.8 percent increases in yeast gene activity, respectively, and that histones associated with yeast heterochromatin are hypomethylated relative to the genome average [Bernstein et al. (2000)]. Speculate on the significance of these findings in terms of nucleosome–DNA interactions and gene activity.
An article entitled 'Nucleosome Positioning at the Replication Fork' states: 'both the 'old' randomly segregated nucleosomes as well as the 'new' assembled histone octamers rapidly position themselves (within seconds) on the newly replicated DNA strands' [Lucchini et al. (2002)]. Given this statement, how would one compare the distribution of nucleosomes and DNA in newly replicated chromatin? How could one experimentally test the distribution of nucleosomes on newly replicated chromosomes?
The following is a diagram of the general structure of the bacteriophage chromosome. Speculate on the mechanism by which it forms a closed ring upon infection of the host cell.
Microsatellites are currently exploited as markers for paternity testing. A sample paternity test is shown in the following table in which ten microsatellite markers were used to test samples from a mother, her child, and an alleged father. The name of the microsatellite locus is given in the left-hand column, and the genotype of each individual is recorded as the number of repeats he or she carries at that locus. For example, at locus D9S302, the mother carries 30 repeats on one of her chromosomes and 31 on the other. In cases where an individual carries the same number of repeats on both chromosomes, only a single number is recorded. (Some of the numbers are followed by a decimal point, for example, 20.2, to indicate a partial repeat in addition to the complete repeats.) Assuming that these markers are inherited in a simple Mendelian fashion, can the alleged father be excluded as the source of the sperm that produced the child? Why or why not? Explain.
At the end of the short arm of human chromosome 16 (16p), several genes associated with disease are present, including thalassemia and polycystic kidney disease. When that region of chromosome 16 was sequenced, gene-coding regions were found to be very close to the telomere-associated sequences. Could there be a possible link between the location of these genes and the presence of the telomere-associated sequences? What further information concerning the disease genes would be useful in your analysis?
