Which of the following can be used to identify an open-reading frame?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
15. Genomes and Genomics
Bioinformatics
Problem 6a
Textbook Question
You are designing algorithms for the bioinformatic prediction of gene sequences. How might algorithms differ for predicting genes in bacterial versus eukaryotic genomic sequence?
 Verified step by step guidance
Verified step by step guidance1
Understand the structural differences between bacterial and eukaryotic genomes: Bacterial genomes are typically smaller, circular, and lack introns, while eukaryotic genomes are larger, linear, and contain introns and exons.
Consider the presence of operons in bacterial genomes: Bacterial genes are often organized into operons, where multiple genes are transcribed together under a single promoter. Algorithms for bacterial gene prediction should account for this feature.
Account for splicing in eukaryotic genomes: Eukaryotic genes contain introns that are spliced out during mRNA processing. Algorithms for eukaryotic gene prediction must identify exon-intron boundaries using splice site signals (e.g., donor and acceptor sites).
Incorporate promoter and regulatory element differences: Bacterial promoters are simpler and have conserved sequences like the -10 and -35 regions, while eukaryotic promoters are more complex and may include TATA boxes and enhancers. Algorithms should be tailored to detect these specific features.
Use codon bias and GC content: Both bacterial and eukaryotic genomes exhibit codon usage bias and characteristic GC content, which can be used to refine gene predictions. However, the patterns differ between the two, so the algorithm must adapt to the organism type.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
2mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Gene Structure Differences
Bacterial genes are typically organized in operons, allowing multiple genes to be transcribed together, while eukaryotic genes are often split by introns and exons, requiring splicing. This structural difference affects how algorithms identify and predict gene sequences, as eukaryotic algorithms must account for splicing and regulatory elements that are less prevalent in bacterial genomes.
Recommended video:
Guided course

Ribosome Structure
Regulatory Elements
Eukaryotic genomes contain complex regulatory elements, such as enhancers and silencers, which influence gene expression. In contrast, bacterial gene regulation is often simpler, relying primarily on promoter regions. Algorithms for eukaryotic gene prediction must incorporate these regulatory sequences to accurately predict gene locations and expression patterns.
Recommended video:
Guided course
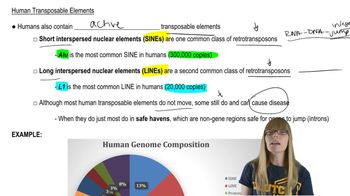
Human Transposable Elements
Sequence Complexity and Size
Eukaryotic genomes are generally larger and more complex than bacterial genomes, containing more repetitive sequences and non-coding DNA. This complexity poses challenges for algorithms, which must efficiently handle larger datasets and distinguish between functional and non-functional sequences, a task that is less demanding in the more streamlined bacterial genomes.
Recommended video:
Guided course

Sanger Sequencing
Related Videos
Related Practice
Multiple Choice
313
views
1
rank


