Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
4. Genetic Mapping and Linkage
Multiple Cross Overs and Interference
Problem 14d
Textbook Question
In Drosophila, a cross was made between females, all expressing the three X-linked recessive traits scute bristles (sc), sable body (s), and vermilion eyes (v)—and wild-type males. In the F₁, all females were wild type, while all males expressed all three mutant traits. The cross was carried to the F₂ generation, and 1000 offspring were counted, with the results shown in the following table.
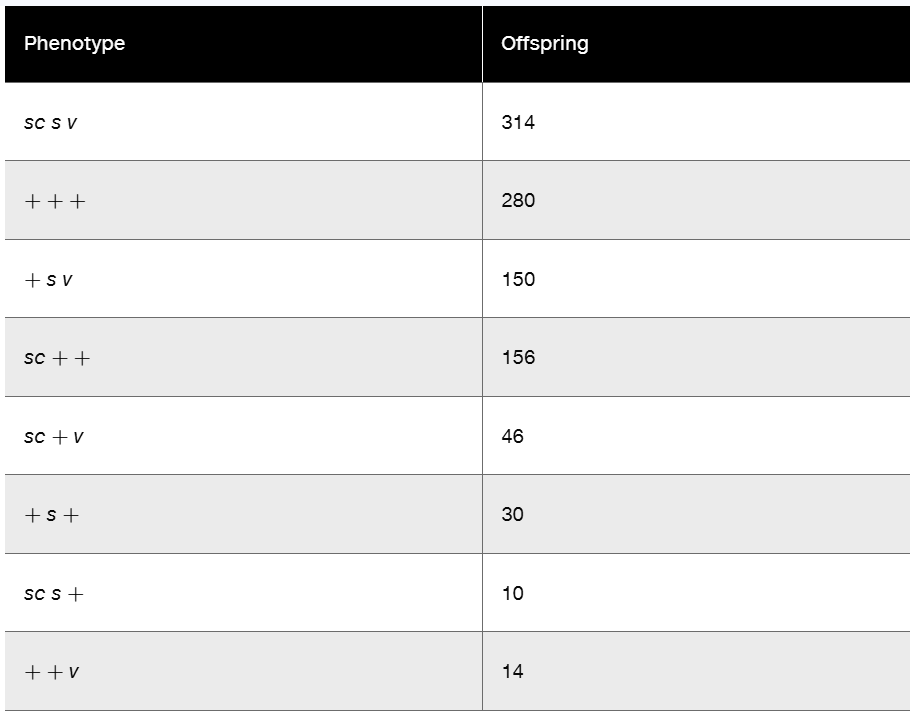
No determination of sex was made in the data. Calculate the coefficient of coincidence. Does it represent positive or negative interference?
 Verified step by step guidance
Verified step by step guidance1
Step 1: Identify the parental and recombinant phenotypes from the table. The parental phenotypes are the most frequent classes, which are 'sc s v' (314 offspring) and '+++ ' (280 offspring). The double crossover classes are the least frequent, which are 'sc + +' (10 offspring) and '++ v' (14 offspring).
Step 2: Calculate the recombination frequencies between the gene pairs. Use the single crossover classes to find the recombination frequency between sc and s, and between s and v. For example, the recombinants between sc and s are the phenotypes where only one of these genes is recombinant (e.g., '+ s v' and 'sc ++'). Calculate the recombination frequency as the sum of these single crossover offspring divided by the total offspring (1000).
Step 3: Calculate the expected number of double crossovers (DCO) assuming independence. Multiply the recombination frequencies of the two intervals to get the expected DCO frequency, then multiply by the total number of offspring to get the expected DCO count.
Step 4: Calculate the coefficient of coincidence (c) using the formula: \(c = \frac{\text{observed DCO}}{\text{expected DCO}}\). The observed DCO is the sum of the two double crossover classes (10 + 14).
Step 5: Determine the interference (I) using the formula: \(I = 1 - c\). If I is positive, it indicates positive interference (fewer double crossovers than expected). If I is negative, it indicates negative interference (more double crossovers than expected).
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
1mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Genetic Linkage and Recombination
Genetic linkage refers to genes located close together on the same chromosome that tend to be inherited together. Recombination occurs during meiosis when crossing over between homologous chromosomes can separate linked genes, producing new allele combinations. The frequency of recombination between genes is used to map their relative positions on chromosomes.
Recommended video:
Guided course

Chi Square and Linkage
Coefficient of Coincidence and Interference
The coefficient of coincidence measures the observed double crossover frequency divided by the expected frequency, indicating how often two crossover events occur together. Interference describes how one crossover event affects the likelihood of another nearby; positive interference reduces double crossovers, while negative interference increases them.
Recommended video:
Guided course

RNA Interference
X-linked Inheritance in Drosophila
In Drosophila, X-linked traits are inherited differently in males and females because males have one X chromosome and females have two. Recessive X-linked traits appear in males if they inherit the mutant allele, while females must inherit two copies. This pattern helps interpret crosses involving X-linked genes and their phenotypic ratios.
Recommended video:
Guided course
X-Inactivation

 6:14m
6:14mWatch next
Master Multiple Cross Overs and Interference with a bite sized video explanation from Kylia
Start learningRelated Videos
Related Practice
Multiple Choice
Which of the following genetic phenomena is most directly influenced by the occurrence of multiple crossovers during meiosis?
29
views
