A female with the following genotype can produce a number of different gametes. Choose the gamete produced if a single crossover has occurred. Genotype = a b + / + + c
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
4. Genetic Mapping and Linkage
Multiple Cross Overs and Interference
Problem 16e
Textbook Question
In a diploid plant species, an F₁ with the genotype Gg Ll Tt is test-crossed to a pure-breeding recessive plant with the genotype gg ll tt. The offspring genotypes are as follows:
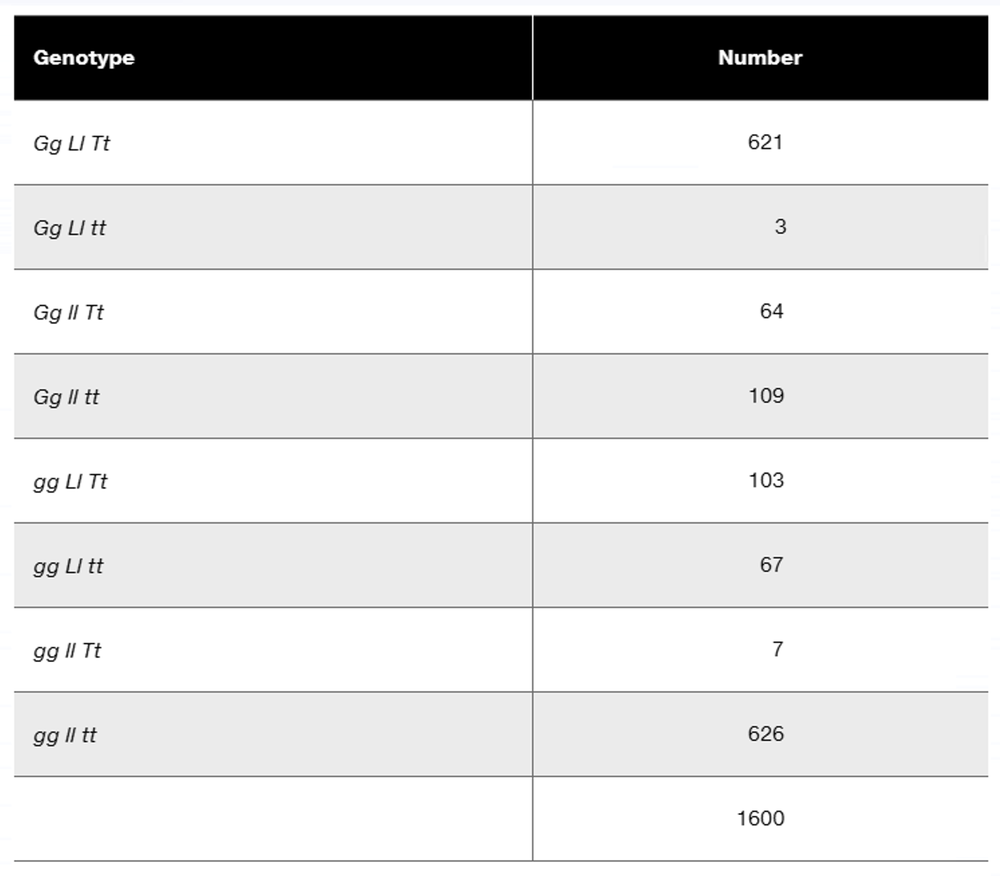
Explain the meaning of this I value.
 Verified step by step guidance
Verified step by step guidance1
Step 1: Understand the problem. The question involves a test cross between an F₁ plant with the genotype Gg Ll Tt and a pure-breeding recessive plant (gg ll tt). The offspring genotypes and their frequencies are provided. The goal is to calculate the I value, which measures interference in genetic recombination.
Step 2: Define interference (I). Interference is a measure of how one crossover event in a region of a chromosome affects the likelihood of another crossover occurring nearby. It is calculated using the formula: I = 1 - (C / E), where C is the observed number of double crossovers, and E is the expected number of double crossovers.
Step 3: Identify the observed double crossovers (C). From the offspring data, the double crossover genotypes are those that result from recombination in both loci. These are the genotypes Gg Ll tt and gg ll Tt. Add their frequencies to find C.
Step 4: Calculate the expected double crossovers (E). To find E, first calculate the recombination frequencies for each gene pair (G-L, L-T, and G-T) using the offspring data. Then, multiply the recombination frequencies for the two relevant gene pairs and the total number of offspring (1600) to estimate E.
Step 5: Calculate the interference (I). Substitute the observed double crossovers (C) and the expected double crossovers (E) into the formula I = 1 - (C / E). This will give you the interference value, which indicates the extent to which crossover events are interfering with each other in this genetic cross.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
2mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Test Cross
A test cross is a genetic cross between an individual with an unknown genotype and a homozygous recessive individual. This method helps determine the genotype of the unknown individual based on the phenotypes of the offspring. In this case, the F₁ plant (Gg Ll Tt) is crossed with a pure-breeding recessive plant (gg ll tt) to reveal the segregation of alleles in the offspring.
Recommended video:
Guided course
Trihybrid Cross
Genotype and Phenotype Ratios
Genotype refers to the genetic constitution of an organism, while phenotype is the observable physical or biochemical characteristics. The offspring's genotypes from the test cross can be analyzed to determine the ratios of different genotypes and phenotypes, which can provide insights into the inheritance patterns and the dominance of alleles in the plant species.
Recommended video:
Guided course

Gamete Genotypes
Chi-Square Test
The Chi-square test is a statistical method used to determine if there is a significant difference between observed and expected frequencies in genetic data. In this context, the I value likely refers to the Chi-square statistic calculated from the observed offspring genotypes compared to the expected ratios based on Mendelian inheritance. A high I value may indicate a deviation from expected ratios, suggesting potential linkage or other genetic interactions.
Recommended video:
Guided course

Chi Square Analysis

 6:14m
6:14mWatch next
Master Multiple Cross Overs and Interference with a bite sized video explanation from Kylia
Start learningRelated Videos
Related Practice
Multiple Choice
705
views
3
rank
4
comments
