Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
18. Molecular Genetic Tools
Genetic Cloning
Problem 17
Textbook Question
The bacteriophage ϕX174 has a single-stranded DNA genome of 5386 bases. During DNA replication, double-stranded forms of the genome are generated. In an effort to create a restriction map of ϕX174, you digest the z-stranded form of the genome with several restriction enzymes and obtain the following results. Draw a map of the ϕX174 genome.
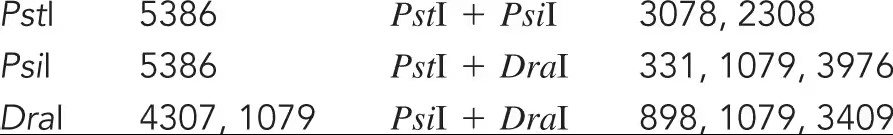
 Verified step by step guidance
Verified step by step guidance1
Step 1: Understand the data provided. The genome of bacteriophage ϕX174 is 5386 bases long. The table shows the fragment sizes obtained after digestion with different restriction enzymes: PstI, PsiI, and DraI, both individually and in combinations. Single enzyme digests give the total fragment sizes, while double digests give overlapping fragment sizes that help locate the cut sites relative to each other.
Step 2: Start by mapping the single enzyme digests. Since PstI and PsiI each cut the genome once (single fragment of 5386 bases), they likely cut at one site each, linearizing the circular genome. DraI cuts twice, producing two fragments of 4307 and 1079 bases, indicating two cut sites.
Step 3: Use the double digest data to determine the relative positions of the cut sites. For example, the PstI + PsiI digest produces fragments of 3078 and 2308 bases. Since PstI and PsiI each cut once, these two fragments sum to 5386 bases, confirming the genome length. This tells us the distance between the PstI and PsiI sites is 2308 bases on one side and 3078 bases on the other.
Step 4: Similarly, analyze the PstI + DraI digest fragments (331, 1079, 3976 bases) and PsiI + DraI digest fragments (898, 1079, 3409 bases). These three fragments sum to 5386 bases, confirming the genome length. Use these fragment sizes to position the DraI sites relative to PstI and PsiI by matching overlapping fragment sizes and distances.
Step 5: Draw the circular genome map by placing the PstI site at an arbitrary position (e.g., 0), then mark the PsiI site 2308 bases away. Next, place the two DraI sites such that the fragment sizes between PstI, PsiI, and DraI sites correspond to the fragment sizes observed in the double digests. This will give a complete restriction map of the ϕX174 genome.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
1mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Restriction Enzyme Digestion and Fragment Analysis
Restriction enzymes cut DNA at specific sequences, producing fragments of varying lengths. By analyzing the sizes of these fragments after digestion, one can infer the locations of enzyme cut sites on the genome. Comparing single and double digests helps determine the relative positions of these sites.
Recommended video:
Guided course

Step 2
Restriction Mapping
Restriction mapping is the process of creating a physical map of a DNA molecule based on the pattern of fragments generated by restriction enzyme digestion. By combining data from single and multiple enzyme digests, the order and distance between restriction sites can be deduced, allowing construction of a genome map.
Recommended video:
Guided course

Mapping with Markers
Double-Stranded DNA Replication of Single-Stranded Genomes
Although ϕX174 has a single-stranded DNA genome, it forms double-stranded replicative forms during replication. Restriction enzymes require double-stranded DNA to cut, so digestion experiments are performed on these replicative forms to analyze the genome structure.
Recommended video:
Guided course

Double Strand Breaks
Related Videos
Related Practice
Textbook Question
A major advance in the 1980s was the development of technology to synthesize short oligonucleotides. This work both facilitated DNA sequencing and led to the advent of the development of PCR. Recently, rapid advances have occurred in the technology to chemically synthesize DNA, and sequences up to 10 kb are now readily produced. As this process becomes more economical, how will it affect the gene-cloning approaches outlined in this chapter? In other words, what types of techniques does this new technology have potential to supplant, and what techniques will not be affected by it?
633
views


