Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
18. Molecular Genetic Tools
Genetic Cloning
Problem 16
Textbook Question
The restriction enzymes XhoI and SalI cut their specific sequences as shown below:

Can the sticky ends created by XhoI and SalI sites be ligated? If yes, can the resulting sequences be cleaved by either XhoI or SalI?
 Verified step by step guidance
Verified step by step guidance1
Step 1: Understand the concept of sticky ends. Sticky ends are single-stranded overhangs created when restriction enzymes cut DNA at specific recognition sites. These overhangs can pair with complementary sequences, allowing DNA fragments to be ligated together.
Step 2: Analyze the sequences cut by XhoI and SalI. XhoI cuts at 5'-CTCGAG-3' and leaves a sticky end with the sequence 5'-TCGA-3'. SalI cuts at 5'-GTCGAC-3' and leaves a sticky end with the sequence 5'-TCGA-3'. Notice that both enzymes produce identical sticky ends (5'-TCGA-3').
Step 3: Determine if ligation is possible. Since the sticky ends produced by XhoI and SalI are identical, they can pair and be ligated together using DNA ligase. This is because complementary base pairing occurs between the overhangs.
Step 4: Examine the resulting sequence after ligation. When the sticky ends are ligated, the resulting sequence will contain the joined DNA fragments. However, the ligation may disrupt the recognition sites for XhoI and SalI, depending on the orientation of the ligated fragments.
Step 5: Assess cleavage by XhoI or SalI. After ligation, check if the recognition sites for XhoI (5'-CTCGAG-3') or SalI (5'-GTCGAC-3') are restored. If the ligation alters the sequence such that the recognition sites are no longer present, the resulting DNA cannot be cleaved by either enzyme.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
2mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
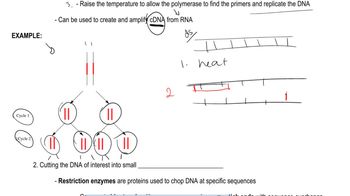
Restriction Enzymes
Restriction enzymes, or restriction endonucleases, are proteins that cut DNA at specific sequences, known as recognition sites. Each enzyme recognizes a unique sequence of nucleotides and cleaves the DNA, often producing 'sticky' or 'blunt' ends. Understanding how these enzymes work is crucial for genetic engineering, as they allow for the manipulation of DNA fragments for cloning or other applications.
Recommended video:
Guided course

Mapping with Markers
Sticky Ends
Sticky ends are short, single-stranded overhangs that are created when restriction enzymes cut DNA in a staggered manner. These overhangs can easily anneal with complementary sequences, facilitating the ligation of different DNA fragments. The ability to ligate DNA fragments with sticky ends is fundamental in recombinant DNA technology, allowing for the creation of new genetic combinations.
Recommended video:
Guided course
Genetic Cloning
Ligation and Cleavage
Ligation is the process of joining two DNA fragments together, typically using the enzyme DNA ligase, which forms covalent bonds between the sugar-phosphate backbones. After ligation, the resulting DNA can be tested for susceptibility to cleavage by the original restriction enzymes. If the ligated sequence contains the recognition sites for either enzyme, it can be cleaved again, which is important for verifying successful cloning or modification.
Recommended video:
Guided course

Post Translational Modifications
Related Videos
Related Practice
Textbook Question
It is often desirable to insert cDNAs into a cloning vector in such a way that all the cDNA clones will have the same orientation with respect to the sequences of the plasmid. This is referred to as directional cloning. Outline how you would directionally clone a cDNA library in the plasmid vector pUC18.
601
views


