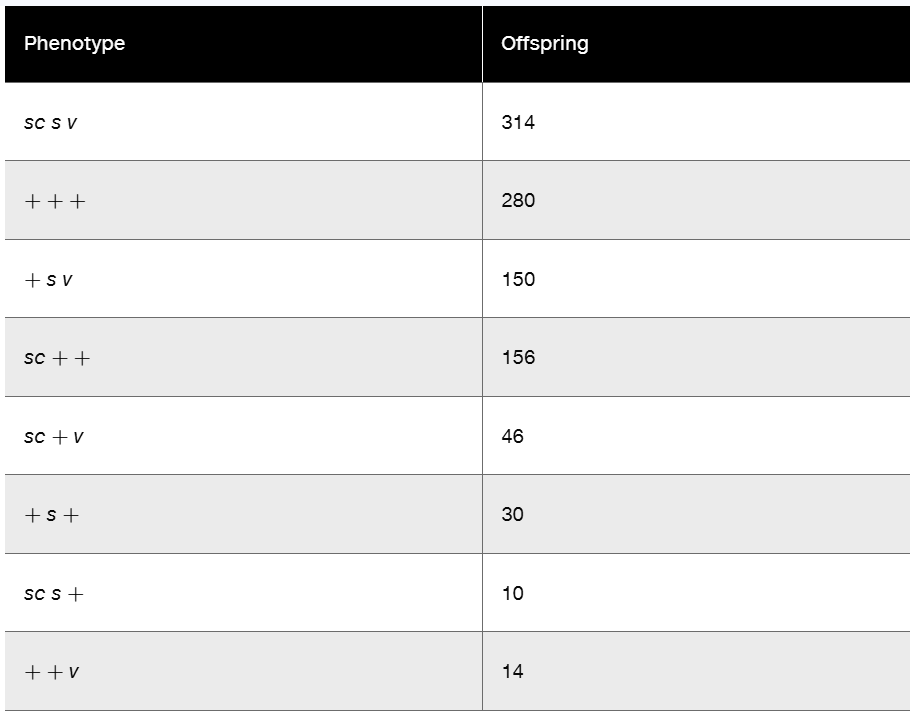
Drosophila melanogaster has one pair of sex chromosomes (XX or XY) and three pairs of autosomes, referred to as chromosomes II, III, and IV. A genetics student discovered a male fly with very short (sh) legs. Using this male, the student was able to establish a pure breeding stock of this mutant and found that it was recessive. She then incorporated the mutant into a stock containing the recessive gene black (b, body color located on chromosome II) and the recessive gene pink (p, eye color located on chromosome III). A female from the homozygous black, pink, short stock was then mated to a wild-type male. The F₁ males of this cross were all wild type and were then backcrossed to the homozygous b, p, sh females. The F₂ results appeared as shown in the following table.
The student repeated the experiment, making the reciprocal cross, with F₁ females backcrossed to homozygous b, p, sh males. She observed that 85 percent of the offspring fell into the given classes, but that 15 percent of the offspring were equally divided among b + p, b + +, + sh p, and + sh + phenotypic males and females. How can these results be explained, and what information can be derived from the data?